A novel feature selection algorithm for identifying hub genes in lung cancer
- PMID: 38066059
- PMCID: PMC10709567
- DOI: 10.1038/s41598-023-48953-1
A novel feature selection algorithm for identifying hub genes in lung cancer
Abstract
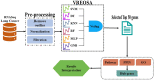
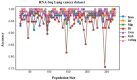
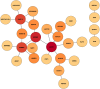
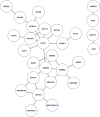
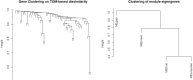
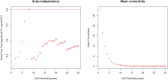
Lung cancer, a life-threatening disease primarily affecting lung tissue, remains a significant contributor to mortality in both developed and developing nations. Accurate biomarker identification is imperative for effective cancer diagnosis and therapeutic strategies. This study introduces the Voting-Based Enhanced Binary Ebola Optimization Search Algorithm (VBEOSA), an innovative ensemble-based approach combining binary optimization and the Ebola optimization search algorithm. VBEOSA harnesses the collective power of the state-of-the-art classification models through soft voting. Moreover, our research applies VBEOSA to an extensive lung cancer gene expression dataset obtained from TCGA, following essential preprocessing steps including outlier detection and removal, data normalization, and filtration. VBEOSA aids in feature selection, leading to the discovery of key hub genes closely associated with lung cancer, validated through comprehensive protein-protein interaction analysis. Notably, our investigation reveals ten significant hub genes-ADRB2, ACTB, ARRB2, GNGT2, ADRB1, ACTG1, ACACA, ATP5A1, ADCY9, and ADRA1B-each demonstrating substantial involvement in the domain of lung cancer. Furthermore, our pathway analysis sheds light on the prominence of strategic pathways such as salivary secretion and the calcium signaling pathway, providing invaluable insights into the intricate molecular mechanisms underpinning lung cancer. We also utilize the weighted gene co-expression network analysis (WGCNA) method to identify gene modules exhibiting strong correlations with clinical attributes associated with lung cancer. Our findings underscore the efficacy of VBEOSA in feature selection and offer profound insights into the multifaceted molecular landscape of lung cancer. Finally, we are confident that this research has the potential to improve diagnostic capabilities and further enrich our understanding of the disease, thus setting the stage for future advancements in the clinical management of lung cancer. The VBEOSA source codes is publicly available at https://github.com/TEHNAN/VBEOSA-A-Novel-Feature-Selection-Algorithm-for-Identifying-hub-Genes-in-Lung-Cancer .
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

