A new subtype of diffuse midline glioma, H3 K27 and BRAF/FGFR1 co-altered: a clinico-radiological and histomolecular characterisation
- PMID: 38066305
- PMCID: PMC10709479
- DOI: 10.1007/s00401-023-02651-4
A new subtype of diffuse midline glioma, H3 K27 and BRAF/FGFR1 co-altered: a clinico-radiological and histomolecular characterisation
Abstract
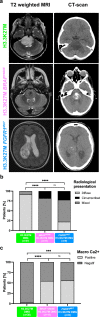
Diffuse midline gliomas (DMG) H3 K27-altered are incurable grade 4 gliomas and represent a major challenge in neuro-oncology. This tumour type is now classified in four subtypes by the 2021 edition of the WHO Classification of the Central Nervous System (CNS) tumours. However, the H3.3-K27M subgroup still appears clinically and molecularly heterogeneous. Recent publications reported that rare patients presenting a co-occurrence of H3.3K27M with BRAF or FGFR1 alterations tended to have a better prognosis. To better study the role of these co-driver alterations, we assembled a large paediatric and adult cohort of 29 tumours H3K27-altered with co-occurring activating mutation in BRAF or FGFR1 as well as 31 previous cases from the literature. We performed a comprehensive histological, radiological, genomic, transcriptomic and DNA methylation analysis. Interestingly, unsupervised t-distributed Stochastic Neighbour Embedding (tSNE) analysis of DNA methylation profiles regrouped BRAFV600E and all but one FGFR1MUT DMG in a unique methylation cluster, distinct from the other DMG subgroups and also from ganglioglioma (GG) or high-grade astrocytoma with piloid features (HGAP). This new DMG subtype harbours atypical radiological and histopathological profiles with calcification and/or a solid tumour component both for BRAFV600E and FGFR1MUT cases. The analyses of a H3.3-K27M BRAFV600E tumour at diagnosis and corresponding in vitro cellular model showed that mutation in H3-3A was the first event in the oncogenesis. Contrary to other DMG, these tumours occur more frequently in the thalamus (70% for BRAFV600E and 58% for FGFR1MUT) and patients have a longer overall survival with a median above three years. In conclusion, DMG, H3 K27 and BRAF/FGFR1 co-altered represent a new subtype of DMG with distinct genotype/phenotype characteristics, which deserve further attention with respect to trial interpretation and patient management.
Keywords: Adult glioma; BRAF-V600E mutation; DNA methylation profiling; FGFR1 mutation; Midline glioma; Paediatric-type high-grade glioma.
© 2023. The Author(s).
Figures




References
-
- Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DTW, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, Radlwimmer B, Højfeldt JW, Truffaux N, Castel D, Schubert S, Ryzhova M, Şeker-Cin H, Gronych J, Johann PD, Stark S, Meyer J, Milde T, Schuhmann M, Ebinger M, Monoranu C-M, Ponnuswami A, Chen S, Jones C, Witt O, Collins VP, von Deimling A, Jabado N, Puget S, Grill J, Helin K, Korshunov A, Lichter P, Monje M, Plass C, Cho Y-J, Pfister SM. Reduced H3K27me3 and DNA Hypomethylation Are Major Drivers of Gene Expression in K27M Mutant Pediatric High-Grade Gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. - DOI - PubMed
-
- Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, Kratz A, Wefers AK, Huang K, Pajtler KW, Schweizer L, Stichel D, Olar A, Engel NW, Lindenberg K, Harter PN, Braczynski AK, Plate KH, Dohmen H, Garvalov BK, Coras R, Hölsken A, Hewer E, Bewerunge-Hudler M, Schick M, Fischer R, Beschorner R, Schittenhelm J, Staszewski O, Wani K, Varlet P, Pages M, Temming P, Lohmann D, Selt F, Witt H, Milde T, Witt O, Aronica E, Giangaspero F, Rushing E, Scheurlen W, Geisenberger C, Rodriguez FJ, Becker A, Preusser M, Haberler C, Bjerkvig R, Cryan J, Farrell M, Deckert M, Hench J, Frank S, Serrano J, Kannan K, Tsirigos A, Brück W, Hofer S, Brehmer S, Seiz-Rosenhagen M, Hänggi D, Hans V, Rozsnoki S, Hansford JR, Kohlhof P, Kristensen BW, Lechner M, Lopes B, Mawrin C, Ketter R, Kulozik A, Khatib Z, Heppner F, Koch A, Jouvet A, Keohane C, Mühleisen H, Mueller W, Pohl U, Prinz M, Benner A, Zapatka M, Gottardo NG, Driever PH, Kramm CM, Müller HL, Rutkowski S, von Hoff K, Frühwald MC, Gnekow A, Fleischhack G, Tippelt S, Calaminus G, Monoranu C-M, Perry A, Jones C, Jacques TS, Radlwimmer B, Gessi M, Pietsch T, Schramm J, Schackert G, Westphal M, Reifenberger G, Wesseling P, Weller M, Collins VP, Blümcke I, Bendszus M, Debus J, Huang A, Jabado N, Northcott PA, Paulus W, Gajjar A, Robinson GW, Taylor MD, Jaunmuktane Z, Ryzhova M, Platten M, Unterberg A, Wick W, Karajannis MA, Mittelbronn M, Acker T, Hartmann C, Aldape K, Schüller U, Buslei R, Lichter P, Kool M, Herold-Mende C, Ellison DW, Hasselblatt M, Snuderl M, Brandner S, Korshunov A, von Deimling A, Pfister SM. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. - DOI - PMC - PubMed
-
- Castel D, Kergrohen T, Tauziède-Espariat A, Mackay A, Ghermaoui S, Lechapt E, Pfister SM, Kramm CM, Boddaert N, Blauwblomme T, Puget S, Beccaria K, Jones C, Jones DTW, Varlet P, Grill J, Debily M-A. Histone H3 wild-type DIPG/DMG overexpressing EZHIP extend the spectrum diffuse midline gliomas with PRC2 inhibition beyond H3–K27M mutation. Acta Neuropathol (Berl) 2020;139:1109–1113. doi: 10.1007/s00401-020-02142-w. - DOI - PubMed
-
- Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, Pagès M, Taylor KR, Saulnier P, Lacroix L, Mackay A, Jones C, Sainte-Rose C, Blauwblomme T, Andreiuolo F, Puget S, Grill J, Varlet P, Debily M-A. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol (Berl) 2015;130:815–827. doi: 10.1007/s00401-015-1478-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

