Pediatric glioma histone H3.3 K27M/G34R mutations drive abnormalities in PML nuclear bodies
- PMID: 38066546
- PMCID: PMC10704828
- DOI: 10.1186/s13059-023-03122-5
Pediatric glioma histone H3.3 K27M/G34R mutations drive abnormalities in PML nuclear bodies
Abstract
Background: Point mutations in histone variant H3.3 (H3.3K27M, H3.3G34R) and the H3.3-specific ATRX/DAXX chaperone complex are frequent events in pediatric gliomas. These H3.3 point mutations affect many chromatin modifications but the exact oncogenic mechanisms are currently unclear. Histone H3.3 is known to localize to nuclear compartments known as promyelocytic leukemia (PML) nuclear bodies, which are frequently mutated and confirmed as oncogenic drivers in acute promyelocytic leukemia.
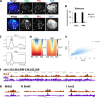
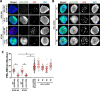
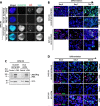
Results: We find that the pediatric glioma-associated H3.3 point mutations disrupt the formation of PML nuclear bodies and this prevents differentiation down glial lineages. Similar to leukemias driven by PML mutations, H3.3-mutated glioma cells are sensitive to drugs that target PML bodies. We also find that point mutations in IDH1/2-which are common events in adult gliomas and myeloid leukemias-also disrupt the formation of PML bodies.
Conclusions: We identify PML as a contributor to oncogenesis in a subset of gliomas and show that targeting PML bodies is effective in treating these H3.3-mutated pediatric gliomas.
Keywords: Arsenic trioxide; Histone variant H3.3; PML bodies; Pediatric glioma.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

