Whole blood transcriptomics identifies subclasses of pediatric septic shock
- PMID: 38066613
- PMCID: PMC10709863
- DOI: 10.1186/s13054-023-04689-y
Whole blood transcriptomics identifies subclasses of pediatric septic shock
Abstract
Background: Sepsis is a highly heterogeneous syndrome, which has hindered the development of effective therapies. This has prompted investigators to develop a precision medicine approach aimed at identifying biologically homogenous subgroups of patients with septic shock and critical illnesses. Transcriptomic analysis can identify subclasses derived from differences in underlying pathophysiological processes that may provide the basis for new targeted therapies. The goal of this study was to elucidate pathophysiological pathways and identify pediatric septic shock subclasses based on whole blood RNA expression profiles.
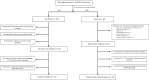
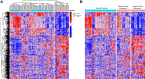
Methods: The subjects were critically ill children with cardiopulmonary failure who were a part of a prospective randomized insulin titration trial to treat hyperglycemia. Genome-wide expression profiling was conducted using RNA sequencing from whole blood samples obtained from 46 children with septic shock and 52 mechanically ventilated noninfected controls without shock. Patients with septic shock were allocated to subclasses based on hierarchical clustering of gene expression profiles, and we then compared clinical characteristics, plasma inflammatory markers, cell compositions using GEDIT, and immune repertoires using Imrep between the two subclasses.
Results: Patients with septic shock depicted alterations in innate and adaptive immune pathways. Among patients with septic shock, we identified two subtypes based on gene expression patterns. Compared with Subclass 2, Subclass 1 was characterized by upregulation of innate immunity pathways and downregulation of adaptive immunity pathways. Subclass 1 had significantly worse clinical outcomes despite the two classes having similar illness severity on initial clinical presentation. Subclass 1 had elevated levels of plasma inflammatory cytokines and endothelial injury biomarkers and demonstrated decreased percentages of CD4 T cells and B cells and less diverse T cell receptor repertoires.
Conclusions: Two subclasses of pediatric septic shock patients were discovered through genome-wide expression profiling based on whole blood RNA sequencing with major biological and clinical differences. Trial Registration This is a secondary analysis of data generated as part of the observational CAF-PINT ancillary of the HALF-PINT study (NCT01565941). Registered March 29, 2012.
Keywords: Adaptive immunity; Gene expression; RNA-Seq; Sepsis; Subclassification.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


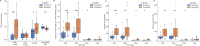
Update of
-
Whole Blood Transcriptomics Identifies Subclasses of Pediatric Septic Shock.Res Sq [Preprint]. 2023 Aug 28:rs.3.rs-3267057. doi: 10.21203/rs.3.rs-3267057/v1. Res Sq. 2023. Update in: Crit Care. 2023 Dec 8;27(1):486. doi: 10.1186/s13054-023-04689-y. PMID: 37693502 Free PMC article. Updated. Preprint.
References
-
- Rogers AJ, Meyer NJ. Precision medicine in critical illness: sepsis and acute respiratory distress syndrome. Precis Pulm Crit Care Sleep Med A Clin Res Guide. 2019 doi: 10.1007/978-3-030-31507-8_18. - DOI
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

