Patterns of Gene Expression, Splicing, and Allele-Specific Expression Vary among Macular Tissues and Clinical Stages of Age-Related Macular Degeneration
- PMID: 38067097
- PMCID: PMC10705168
- DOI: 10.3390/cells12232668
Patterns of Gene Expression, Splicing, and Allele-Specific Expression Vary among Macular Tissues and Clinical Stages of Age-Related Macular Degeneration
Abstract
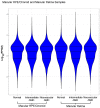
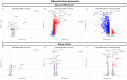
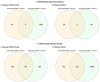
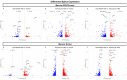
Age-related macular degeneration (AMD) is a leading cause of blindness, and elucidating its underlying disease mechanisms is vital to the development of appropriate therapeutics. We identified differentially expressed genes (DEGs) and differentially spliced genes (DSGs) across the clinical stages of AMD in disease-affected tissue, the macular retina pigment epithelium (RPE)/choroid and the macular neural retina within the same eye. We utilized 27 deeply phenotyped donor eyes (recovered within a 6 h postmortem interval time) from Caucasian donors (60-94 years) using a standardized published protocol. Significant findings were then validated in an independent set of well-characterized donor eyes (n = 85). There was limited overlap between DEGs and DSGs, suggesting distinct mechanisms at play in AMD pathophysiology. A greater number of previously reported AMD loci overlapped with DSGs compared to DEGs between disease states, and no DEG overlap with previously reported loci was found in the macular retina between disease states. Additionally, we explored allele-specific expression (ASE) in coding regions of previously reported AMD risk loci, uncovering a significant imbalance in C3 rs2230199 and CFH rs1061170 in the macular RPE/choroid for normal eyes and intermediate AMD (iAMD), and for CFH rs1061147 in the macular RPE/choroid for normal eyes and iAMD, and separately neovascular AMD (NEO). Only significant DEGs/DSGs from the macular RPE/choroid were found to overlap between disease states. STAT1, validated between the iAMD vs. normal comparison, and AGTPBP1, BBS5, CERKL, FGFBP2, KIFC3, RORα, and ZNF292, validated between the NEO vs. normal comparison, revealed an intricate regulatory network with transcription factors and miRNAs identifying potential upstream and downstream regulators. Findings regarding the complement genes C3 and CFH suggest that coding variants at these loci may influence AMD development via an imbalance of gene expression in a tissue-specific manner. Our study provides crucial insights into the multifaceted genomic underpinnings of AMD (i.e., tissue-specific gene expression changes, potential splice variation, and allelic imbalance), which may open new avenues for AMD diagnostics and therapies specific to iAMD and NEO.
Keywords: age-related macular degeneration (AMD); allele-specific expression (ASE); and AMD therapies; intermediate AMD (iAMD); macular retina; macular retinal pigment epithelium (RPE)/choroid; microRNAs (miRNAs); neovascular AMD (NEO); splicing; tissue-specific gene expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ung C., Lains I., Miller J.W., Kim I.K. Current Management of Age-Related Macular Degeneration. In: Chew E.Y., Swaroop A., editors. Age-Related Macular Degeneration. Springer International Publishing; Cham, Switzerland: 2021. pp. 295–314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

