Altered Regulation of the Glucose Transporter GLUT3 in PRDX1 Null Cells Caused Hypersensitivity to Arsenite
- PMID: 38067110
- PMCID: PMC10705171
- DOI: 10.3390/cells12232682
Altered Regulation of the Glucose Transporter GLUT3 in PRDX1 Null Cells Caused Hypersensitivity to Arsenite
Abstract
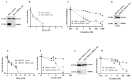
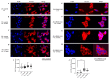
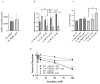
Targeting tumour metabolism through glucose transporters is an attractive approach. However, the role these transporters play through interaction with other signalling proteins is not yet defined. The glucose transporter SLC2A3 (GLUT3) is a member of the solute carrier transporter proteins. GLUT3 has a high affinity for D-glucose and regulates glucose uptake in the neurons, as well as other tissues. Herein, we show that GLUT3 is involved in the uptake of arsenite, and its level is regulated by peroxiredoxin 1 (PRDX1). In the absence of PRDX1, GLUT3 mRNA and protein expression levels are low, but they are increased upon arsenite treatment, correlating with an increased uptake of glucose. The downregulation of GLUT3 by siRNA or deletion of the gene by CRISPR cas-9 confers resistance to arsenite. Additionally, the overexpression of GLUT3 sensitises the cells to arsenite. We further show that GLUT3 interacts with PRDX1, and it forms nuclear foci, which are redistributed upon arsenite exposure, as revealed by immunofluorescence analysis. We propose that GLUT3 plays a role in mediating the uptake of arsenite into cells, and its homeostatic and redox states are tightly regulated by PRDX1. As such, GLUT3 and PRDX1 are likely to be novel targets for arsenite-based cancer therapy.
Keywords: GLUT3 redox state; PRDX1; SLC2A3; arsenite sensitivity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Regulation of GLUT3 and glucose uptake by the cAMP signalling pathway in the breast cancer cell line ZR-75.J Cell Physiol. 2008 Jan;214(1):110-6. doi: 10.1002/jcp.21166. J Cell Physiol. 2008. PMID: 17559076
-
Expression of hypoxia-related glucose transporters GLUT1 and GLUT3 in benign, malignant and non-neoplastic thyroid lesions.Mol Med Rep. 2012 Sep;6(3):601-6. doi: 10.3892/mmr.2012.969. Epub 2012 Jun 27. Mol Med Rep. 2012. PMID: 22752218
-
CREB regulates the expression of neuronal glucose transporter 3: a possible mechanism related to impaired brain glucose uptake in Alzheimer's disease.Nucleic Acids Res. 2013 Mar 1;41(5):3240-56. doi: 10.1093/nar/gks1227. Epub 2013 Jan 22. Nucleic Acids Res. 2013. PMID: 23341039 Free PMC article.
-
Glucose transporter 3 in neuronal glucose metabolism: Health and diseases.Metabolism. 2021 Oct;123:154869. doi: 10.1016/j.metabol.2021.154869. Epub 2021 Aug 21. Metabolism. 2021. PMID: 34425073 Review.
-
Cellular effects and clinical implications of SLC2A3 copy number variation.J Cell Physiol. 2020 Dec;235(12):9021-9036. doi: 10.1002/jcp.29753. Epub 2020 May 5. J Cell Physiol. 2020. PMID: 32372501 Review.
Cited by
-
Proteomic-miRNA Biomics Profile Reveals 2D Cultures of Human iPSC-Derived Neural Progenitor Cells More Sensitive than 3D Spheroid System Against the Experimental Exposure to Arsenic.Mol Neurobiol. 2024 Aug;61(8):5754-5770. doi: 10.1007/s12035-024-03924-z. Epub 2024 Jan 16. Mol Neurobiol. 2024. PMID: 38228842
-
PRDX1 protects ATM from arsenite-induced proteotoxicity and maintains its stability during DNA damage signaling.Oncotarget. 2025 May 19;16:362-378. doi: 10.18632/oncotarget.28720. Oncotarget. 2025. PMID: 40387816 Free PMC article.
-
Malignant features related PRDX1 associated with osimertinib sensitivity of EGFR-mutant lung adenocarcinoma.Int J Med Sci. 2025 Mar 31;22(9):2040-2058. doi: 10.7150/ijms.107255. eCollection 2025. Int J Med Sci. 2025. PMID: 40303499 Free PMC article.
-
GLUT3 enhances chemosensitivity in glioblastoma by transporting temozolomide and capecitabine.Cell Death Discov. 2025 Aug 14;11(1):382. doi: 10.1038/s41420-025-02664-w. Cell Death Discov. 2025. PMID: 40813374 Free PMC article.
References
-
- Zhang H.N., Yang L., Ling J.Y., Czajkowsky D.M., Wang J.F., Zhang X.W., Zhou Y.M., Ge F., Yang M.K., Xiong Q., et al. Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc. Natl. Acad. Sci. USA. 2015;112:15084–15089. doi: 10.1073/pnas.1521316112. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

