Cell Adhesion at the Tight Junctions: New Aspects and New Functions
- PMID: 38067129
- PMCID: PMC10706136
- DOI: 10.3390/cells12232701
Cell Adhesion at the Tight Junctions: New Aspects and New Functions
Abstract
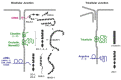
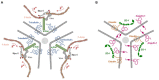
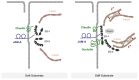
Tight junctions (TJ) are cell-cell adhesive structures that define the permeability of barrier-forming epithelia and endothelia. In contrast to this seemingly static function, TJs display a surprisingly high molecular complexity and unexpected dynamic regulation, which allows the TJs to maintain a barrier in the presence of physiological forces and in response to perturbations. Cell-cell adhesion receptors play key roles during the dynamic regulation of TJs. They connect individual cells within cellular sheets and link sites of cell-cell contacts to the underlying actin cytoskeleton. Recent findings support the roles of adhesion receptors in transmitting mechanical forces and promoting phase separation. In this review, we discuss the newly discovered functions of cell adhesion receptors localized at the TJs and their role in the regulation of the barrier function.
Keywords: JAM-A; cell–cell adhesion; force sensing; junctional adhesion molecule; phase separation; tight junction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Human junction adhesion molecule regulates tight junction resealing in epithelia.J Cell Sci. 2000 Jul;113 ( Pt 13):2363-74. doi: 10.1242/jcs.113.13.2363. J Cell Sci. 2000. PMID: 10852816
-
Nectin and junctional adhesion molecule are critical cell adhesion molecules for the apico-basal alignment of adherens and tight junctions in epithelial cells.Genes Cells. 2013 Nov;18(11):985-98. doi: 10.1111/gtc.12091. Epub 2013 Sep 24. Genes Cells. 2013. PMID: 24112238
-
JAM-A associates with ZO-2, afadin, and PDZ-GEF1 to activate Rap2c and regulate epithelial barrier function.Mol Biol Cell. 2013 Sep;24(18):2849-60. doi: 10.1091/mbc.E13-06-0298. Epub 2013 Jul 24. Mol Biol Cell. 2013. PMID: 23885123 Free PMC article.
-
Apical cytoskeletons and junctional complexes as a combined system in epithelial cell sheets.Ann N Y Acad Sci. 2017 Oct;1405(1):32-43. doi: 10.1111/nyas.13432. Epub 2017 Aug 1. Ann N Y Acad Sci. 2017. PMID: 28763830 Review.
-
Tight Junction Structure and Function Revisited.Trends Cell Biol. 2020 Oct;30(10):805-817. doi: 10.1016/j.tcb.2020.08.004. Epub 2020 Sep 2. Trends Cell Biol. 2020. PMID: 32891490 Review.
Cited by
-
Quercetin Improves Barrier Properties in Porcine Small Intestine but Not in Peyer's Patches.Int J Mol Sci. 2024 Jan 26;25(3):1530. doi: 10.3390/ijms25031530. Int J Mol Sci. 2024. PMID: 38338808 Free PMC article.
-
Looking Inside of the Intestinal Permeability Regulation by Protein-Derivatives from Bovine Milk.Mol Nutr Food Res. 2024 Nov;68(22):e2400384. doi: 10.1002/mnfr.202400384. Epub 2024 Nov 12. Mol Nutr Food Res. 2024. PMID: 39530631 Free PMC article. Review.
-
Regulation of Vascular Injury and Repair by P21-Activated Kinase 1 and P21-Activated Kinase 2: Therapeutic Potential and Challenges.Biomolecules. 2024 Dec 13;14(12):1596. doi: 10.3390/biom14121596. Biomolecules. 2024. PMID: 39766303 Free PMC article. Review.
-
Hsa-miR-31-3p targets CLDN8 to compromise skin barrier integrity in psoriasis.Biochem Biophys Rep. 2025 Mar 13;42:101976. doi: 10.1016/j.bbrep.2025.101976. eCollection 2025 Jun. Biochem Biophys Rep. 2025. PMID: 40160514 Free PMC article.
-
Panose prevents acute-on-chronic liver failure by reducing bacterial infection in mice.J Clin Invest. 2025 Jun 3;135(14):e184653. doi: 10.1172/JCI184653. eCollection 2025 Jul 15. J Clin Invest. 2025. PMID: 40478727 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

