Spatial Transcriptomics Reveals Signatures of Histopathological Changes in Muscular Sarcoidosis
- PMID: 38067175
- PMCID: PMC10706822
- DOI: 10.3390/cells12232747
Spatial Transcriptomics Reveals Signatures of Histopathological Changes in Muscular Sarcoidosis
Abstract
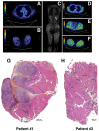
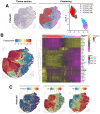
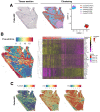
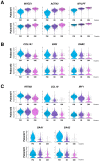
Sarcoidosis is a multisystemic disease characterized by non-caseating granuloma infiltrating various organs. The form with symptomatic muscular involvement is called muscular sarcoidosis. The impact of immune cells composing the granuloma on the skeletal muscle is misunderstood. Here, we investigated the granuloma-skeletal muscle interactions through spatial transcriptomics on two patients affected by muscular sarcoidosis. Five major transcriptomic clusters corresponding to perigranuloma, granuloma, and three successive muscle tissue areas (proximal, intermediate, and distal) around the granuloma were identified. Analyses revealed upregulated pathways in the granuloma corresponding to the activation of T-lymphocytes and monocytes/macrophages cytokines, the upregulation of extracellular matrix signatures, and the induction of the TGF-β signaling in the perigranuloma. A comparison between the proximal and distal muscles to the granuloma revealed an inverse correlation between the distance to the granuloma and the upregulation of cellular response to interferon-γ/α, TNF-α, IL-1,4,6, fibroblast proliferation, epithelial to mesenchymal cell transition, and the downregulation of muscle gene expression. These data shed light on the intercommunications between granulomas and the muscle tissue and provide pathophysiological mechanisms by showing that granuloma immune cells have a direct impact on proximal muscle tissue by promoting its progressive replacement by fibrosis via the expression of pro-inflammatory and profibrosing signatures. These data could possibly explain the evolution towards a state of disability for some patients.
Keywords: Visium; fibrosis; granuloma; muscular sarcoidosis; skeletal muscle; spatial transcriptomic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

