Role of Phosphodiesterase 1 in the Regulation of Real-Time cGMP Levels and Contractility in Adult Mouse Cardiomyocytes
- PMID: 38067187
- PMCID: PMC10706287
- DOI: 10.3390/cells12232759
Role of Phosphodiesterase 1 in the Regulation of Real-Time cGMP Levels and Contractility in Adult Mouse Cardiomyocytes
Abstract
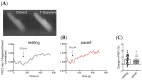
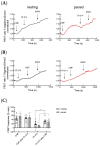
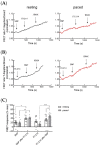
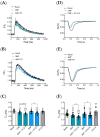
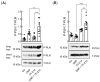
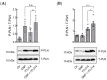
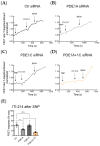
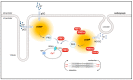
In mouse cardiomyocytes, the expression of two subfamilies of the calcium/calmodulin-regulated cyclic nucleotide phosphodiesterase 1 (PDE1)-PDE1A and PDE1C-has been reported. PDE1C was found to be the major subfamily in the human heart. It is a dual substrate PDE and can hydrolyze both 3',5'-cyclic adenosine monophosphate (cAMP) and 3',5'-cyclic guanosine monophosphate (cGMP). Previously, it has been reported that the PDE1 inhibitor ITI-214 shows positive inotropic effects in heart failure patients which were largely attributed to the cAMP-dependent protein kinase (PKA) signaling. However, the role of PDE1 in the regulation of cardiac cGMP has not been directly addressed. Here, we studied the effect of PDE1 inhibition on cGMP levels in adult mouse ventricular cardiomyocytes using a highly sensitive fluorescent biosensor based on Förster resonance energy transfer (FRET). Live-cell imaging in paced and resting cardiomyocytes showed an increase in cGMP after PDE1 inhibition with ITI-214. Furthermore, PDE1 inhibition and PDE1A knockdown amplified the cGMP-FRET responses to the nitric oxide (NO)-donor sodium nitroprusside (SNP) but not to the C-type natriuretic peptide (CNP), indicating a specific role of PDE1 in the regulation of the NO-sensitive guanylyl cyclase (NO-GC)-regulated cGMP microdomain. ITI-214, in combination with CNP or SNP, showed a positive lusitropic effect, improving the relaxation of isolated myocytes. Immunoblot analysis revealed increased phospholamban (PLN) phosphorylation at Ser-16 in cells treated with a combination of SNP and PDE1 inhibitor but not with SNP alone. Our findings reveal a previously unreported role of PDE1 in the regulation of the NO-GC/cGMP microdomain and mouse ventricular myocyte contractility. Since PDE1 serves as a cGMP degrading PDE in cardiomyocytes and has the highest hydrolytic activities, it can be expected that PDE1 inhibition might be beneficial in combination with cGMP-elevating drugs for the treatment of cardiac diseases.
Keywords: FRET; PDE1; cGMP; cardiomyocyte.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

