Diverse Transcriptome Responses to Salinity Change in Atlantic Cod Subpopulations
- PMID: 38067188
- PMCID: PMC10706248
- DOI: 10.3390/cells12232760
Diverse Transcriptome Responses to Salinity Change in Atlantic Cod Subpopulations
Abstract
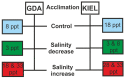
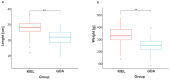
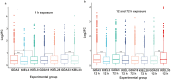
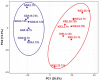
Adaptation to environmental variation caused by global climate change is a significant aspect of fisheries management and ecology. A reduction in ocean salinity is visible in near-shore areas, especially in the Baltic Sea, where it is affecting the Atlantic cod population. Cod is one of the most significant teleost species, with high ecological and economical value worldwide. The population of cod in the Baltic Sea has been traditionally divided into two subpopulations (western and eastern) existing in higher- and lower-salinity waters, respectively. In recent decades, both Baltic cod subpopulations have declined massively. One of the reasons for the poor condition of cod in the Baltic Sea is environmental factors, including salinity. Thus, in this study, an oligonucleotide microarray was applied to explore differences between Baltic cod subpopulations in response to salinity fluctuations. For this purpose, an exposure experiment was conducted consisting of salinity elevation and reduction, and gene expression was measured in gill tissue. We found 400 differentially expressed genes (DEGs) involved in the immune response, metabolism, programmed cell death, cytoskeleton, and extracellular matrix that showed a subpopulation-dependent pattern. These findings indicate that osmoregulation in Baltic cod is a complex process, and that western and eastern Baltic cod subpopulations respond differently to salinity changes.
Keywords: Gadus morhua; gene expression; gill tissue; microarray; osmoregulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Transcriptome analysis of gill tissue of Atlantic cod Gadus morhua L. from the Baltic Sea.Mar Genomics. 2015 Oct;23:37-40. doi: 10.1016/j.margen.2015.04.005. Epub 2015 Apr 24. Mar Genomics. 2015. PMID: 25913867
-
SNP genotyping reveals substructuring in weakly differentiated populations of Atlantic cod (Gadus morhua) from diverse environments in the Baltic Sea.Sci Rep. 2020 Jun 16;10(1):9738. doi: 10.1038/s41598-020-66518-4. Sci Rep. 2020. PMID: 32546719 Free PMC article.
-
Adaptation to Low Salinity Promotes Genomic Divergence in Atlantic Cod (Gadus morhua L.).Genome Biol Evol. 2015 May 20;7(6):1644-63. doi: 10.1093/gbe/evv093. Genome Biol Evol. 2015. PMID: 25994933 Free PMC article.
-
Use of existing hydrographic infrastructure to forecast the environmental spawning conditions for Eastern Baltic cod.PLoS One. 2018 May 16;13(5):e0196477. doi: 10.1371/journal.pone.0196477. eCollection 2018. PLoS One. 2018. PMID: 29768443 Free PMC article.
-
Analysing migrations of Atlantic cod Gadus morhua in the north-east Atlantic Ocean: then, now and the future.J Fish Biol. 2013 Mar;82(3):741-63. doi: 10.1111/jfb.12043. J Fish Biol. 2013. PMID: 23464542 Review.
References
-
- Gutiérrez-Cánovas C., Millán A., Velasco J., Vaughan I.P., Ormerod S.J. Contrasting effects of natural and anthropogenic stressors on beta diversity in river organisms. Glob. Ecol. Biogeogr. 2013;22:796–805. doi: 10.1111/geb.12060. - DOI
-
- Lehmann A., Myrberg K., Post P., Chubarenko I., Dailidiene I., Hinrichsen H.-H., Hüssy K., Liblik T., Meier H.E.M., Lips U., et al. Salinity dynamics of the Baltic Sea. Earth Syst. Dyn. 2022;13:373–392. doi: 10.5194/esd-13-373-2022. - DOI
-
- Meier H.E.M., Kjellström E., Graham L.P. Estimating uncertainties of projected Baltic Sea salinity in the late 21st century. Geophys. Res. Lett. 2006;33:L15705. doi: 10.1029/2006GL026488. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

