The MYC-Regulated RNA-Binding Proteins hnRNPC and LARP1 Are Drivers of Multiple Myeloma Cell Growth and Disease Progression and Negatively Predict Patient Survival
- PMID: 38067212
- PMCID: PMC10705803
- DOI: 10.3390/cancers15235508
The MYC-Regulated RNA-Binding Proteins hnRNPC and LARP1 Are Drivers of Multiple Myeloma Cell Growth and Disease Progression and Negatively Predict Patient Survival
Abstract
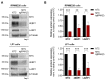
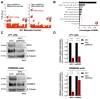
Multiple myeloma (MM) is a malignant plasma cell disorder in which the MYC oncogene is frequently dysregulated. Due to its central role, MYC has been proposed as a drug target; however, the development of a clinically applicable molecule modulating MYC activity remains an unmet challenge. Consequently, an alternative is the development of therapeutic options targeting proteins located downstream of MYC. Therefore, we aimed to identify undescribed MYC-target proteins in MM cells using Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) and mass spectrometry. We revealed a cluster of proteins associated with the regulation of translation initiation. Herein, the RNA-binding proteins Heterogeneous Nuclear Ribonucleoprotein C (hnRNPC) and La Ribonucleoprotein 1 (LARP1) were predominantly downregulated upon MYC depletion. CRISPR-mediated knockout of either hnRNPC or LARP1 in conjunction with redundant LARP family proteins resulted in a proliferative disadvantage for MM cells. Moreover, high expression levels of these proteins correlate with high MYC expression and with poor survival and disease progression in MM patients. In conclusion, our study provides valuable insights into MYC's role in translation initiation by identifying hnRNPC and LARP1 as proliferation drivers of MM cells and as both predictive factors for survival and disease progression in MM patients.
Keywords: La Ribonucleoprotein 1 (LARP1); MYC target pathways; RNA-binding proteins (RBPs); heterogeneous nuclear ribonucleoprotein C (hnRNPC); multiple myeloma; translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Circular RNA circ-FIRRE interacts with HNRNPC to promote esophageal squamous cell carcinoma progression by stabilizing GLI2 mRNA.Cancer Sci. 2023 Sep;114(9):3608-3622. doi: 10.1111/cas.15899. Epub 2023 Jul 7. Cancer Sci. 2023. PMID: 37417427 Free PMC article.
-
S-adenosylmethionine inhibits la ribonucleoprotein domain family member 1 in murine liver and human liver cancer cells.Hepatology. 2022 Feb;75(2):280-296. doi: 10.1002/hep.32130. Epub 2021 Dec 18. Hepatology. 2022. PMID: 34449924 Free PMC article.
-
HNRNPC downregulation inhibits IL-6/STAT3-mediated HCC metastasis by decreasing HIF1A expression.Cancer Sci. 2022 Oct;113(10):3347-3361. doi: 10.1111/cas.15494. Epub 2022 Aug 12. Cancer Sci. 2022. PMID: 35848884 Free PMC article.
-
LARP1 on TOP of ribosome production.Wiley Interdiscip Rev RNA. 2018 Sep;9(5):e1480. doi: 10.1002/wrna.1480. Epub 2018 May 2. Wiley Interdiscip Rev RNA. 2018. PMID: 29722158 Free PMC article. Review.
-
The La-Related Proteins, a Family with Connections to Cancer.Biomolecules. 2015 Oct 16;5(4):2701-22. doi: 10.3390/biom5042701. Biomolecules. 2015. PMID: 26501340 Free PMC article. Review.
Cited by
-
Bioinformatics identification and validation of m6A/m1A/m5C/m7G/ac4 C-modified genes in oral squamous cell carcinoma.BMC Cancer. 2025 Jul 1;25(1):1055. doi: 10.1186/s12885-025-14216-7. BMC Cancer. 2025. PMID: 40597017 Free PMC article.
-
Telocinobufagin suppresses malignant metastasis of undifferentiated thyroid carcinoma via modulation of the LARP1-mTOR pathway.Kaohsiung J Med Sci. 2025 Mar;41(3):e12934. doi: 10.1002/kjm2.12934. Epub 2025 Jan 9. Kaohsiung J Med Sci. 2025. PMID: 39786317 Free PMC article.
-
Molecular heterogeneity and MYC dysregulation in triple-negative breast cancer: genomic advances and therapeutic implications.3 Biotech. 2025 Jan;15(1):33. doi: 10.1007/s13205-024-04195-0. Epub 2025 Jan 5. 3 Biotech. 2025. PMID: 39777154 Review.
-
Mechanism of the N6-methyladenosine reader heterogeneous nuclear ribonucleoprotein C facilitating immune escape in thyroid cancer by stabilizing programmed death ligand 1.Cytotechnology. 2025 Jun;77(3):96. doi: 10.1007/s10616-025-00755-1. Epub 2025 May 6. Cytotechnology. 2025. PMID: 40342628
References
-
- Hansen D.K., Sidana S., Peres L.C., Colin Leitzinger C., Shune L., Shrewsbury A., Gonzalez R., Sborov D.W., Wagner C., Dima D., et al. Idecabtagene Vicleucel for Relapsed/Refractory Multiple Myeloma: Real-World Experience from the Myeloma CAR T Consortium. J. Clin. Oncol. 2023;41:2087–2097. doi: 10.1200/JCO.22.01365. - DOI - PMC - PubMed
-
- Chng W.J., Huang G.F., Chung T.H., Ng S.B., Gonzalez-Paz N., Troska-Price T., Mulligan G., Chesi M., Bergsagel P.L., Fonseca R. Clinical and biological implications of MYC activation: A common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25:1026–1035. doi: 10.1038/leu.2011.53. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

