MiRNAs in Alcohol-Related Liver Diseases and Hepatocellular Carcinoma: A Step toward New Therapeutic Approaches?
- PMID: 38067261
- PMCID: PMC10705678
- DOI: 10.3390/cancers15235557
MiRNAs in Alcohol-Related Liver Diseases and Hepatocellular Carcinoma: A Step toward New Therapeutic Approaches?
Abstract
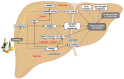
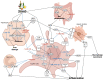
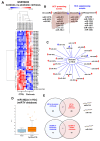
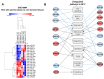
Alcohol-related Liver Disease (ALD) is the primary cause of chronic liver disorders and hepatocellular carcinoma (HCC) development in developed countries and thus represents a major public health concern. Unfortunately, few therapeutic options are available for ALD and HCC, except liver transplantation or tumor resection for HCC. Deciphering the molecular mechanisms underlying the development of these diseases is therefore of major importance to identify early biomarkers and to design efficient therapeutic options. Increasing evidence indicate that epigenetic alterations play a central role in the development of ALD and HCC. Among them, microRNA importantly contribute to the development of this disease by controlling the expression of several genes involved in hepatic metabolism, inflammation, fibrosis, and carcinogenesis at the post-transcriptional level. In this review, we discuss the current knowledge about miRNAs' functions in the different stages of ALD and their role in the progression toward carcinogenesis. We highlight that each stage of ALD is associated with deregulated miRNAs involved in hepatic carcinogenesis, and thus represent HCC-priming miRNAs. By using in silico approaches, we have uncovered new miRNAs potentially involved in HCC. Finally, we discuss the therapeutic potential of targeting miRNAs for the treatment of these diseases.
Keywords: alcohol-related liver disease; hepatocellular carcinoma; miRNAs-based therapeutics; microRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

