Identification of Tumor-Suppressive miR-139-3p- Regulated Genes: TRIP13 as a Therapeutic Target in Lung Adenocarcinoma
- PMID: 38067275
- PMCID: PMC10705761
- DOI: 10.3390/cancers15235571
Identification of Tumor-Suppressive miR-139-3p- Regulated Genes: TRIP13 as a Therapeutic Target in Lung Adenocarcinoma
Abstract
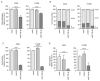
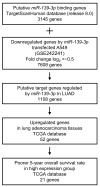
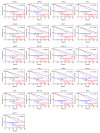
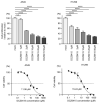
Analyses of our microRNA (miRNA) expression signature combined with The Cancer Genome Atlas (TCGA) data revealed that both strands of pre-miR-139 (miR-139-5p, the guide strand, and miR-139-3p, the passenger strand) are significantly downregulated in lung adenocarcinoma (LUAD) clinical specimens. Functional analyses of LUAD cells ectopically expressing miR-139-3p showed significant suppression of their aggressiveness (e.g., cancer cell proliferation, migration, and invasion). The involvement of the passenger strand, miR-139-3p, in LUAD pathogenesis, is an interesting finding contributing to the elucidation of unknown molecular networks in LUAD. Of 1108 genes identified as miR-139-3p targets in LUAD cells, 21 were significantly upregulated in LUAD tissues according to TCGA analysis, and their high expression negatively affected the prognosis of LUAD patients. We focused on thyroid hormone receptor interactor 13 (TRIP13) and investigated its cancer-promoting functions in LUAD cells. Luciferase assays showed that miR-139-3p directly regulated TRIP13. siRNA-mediated TRIP13 knockdown and TRIP13 inhibition by a specific inhibitor (DCZ0415) attenuated the malignant transformation of LUAD cells. Interestingly, when used in combination with anticancer drugs (cisplatin and carboplatin), DCZ0415 exerted synergistic effects on cell proliferation suppression. Identifying the molecular pathways regulated by tumor-suppressive miRNAs (including passenger strands) may aid in the discovery of diagnostic markers and therapeutic targets for LUAD.
Keywords: TRIP13; lung adenocarcinoma; miR-139-3p; microRNA; passenger strand.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Thai A.A., Solomon B.J., Sequist L.V., Gainor J.F., Heist R.S. Lung cancer. Lancet. 2021;398:535–554. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

