Characteristics of ABCC4 and ABCG2 High Expression Subpopulations in CRC-A New Opportunity to Predict Therapy Response
- PMID: 38067326
- PMCID: PMC10705186
- DOI: 10.3390/cancers15235623
Characteristics of ABCC4 and ABCG2 High Expression Subpopulations in CRC-A New Opportunity to Predict Therapy Response
Abstract
Background: Our previous findings proved that ABCC4 and ABCG2 proteins present much more complex roles in colorectal cancer (CRC) than typically cancer-associated functions as drug exporters. Our objective was to evaluate their predictive/diagnostic potential.
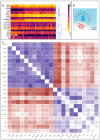
Methods: CRC patients' transcriptomic data from the Gene Expression Omnibus database (GSE18105, GSE21510 and GSE41568) were discriminated into two subpopulations presenting either high expression levels of ABCC4 (ABCC4 High) or ABCG2 (ABCG2 High). Subpopulations were analysed using various bioinformatical tools and platforms (KEEG, Gene Ontology, FunRich v3.1.3, TIMER2.0 and STRING 12.0).
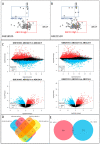
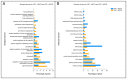
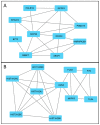
Results: The analysed subpopulations present different gene expression patterns. The protein-protein interaction network of subpopulation-specific genes revealed the top hub proteins in ABCC4 High: RPS27A, SRSF1, DDX3X, BPTF, RBBP7, POLR1B, HNRNPA2B1, PSMD14, NOP58 and EIF2S3 and in ABCG2 High: MAPK3, HIST2H2BE, LMNA, HIST1H2BD, HIST1H2BK, HIST1H2AC, FYN, TLR4, FLNA and HIST1H2AJ. Additionally, our multi-omics analysis proved that the ABCC4 expression correlates with substantially increased tumour-associated macrophage infiltration and sensitivity to FOLFOX treatment.
Conclusions: ABCC4 and ABCG2 may be used to distinguish CRC subpopulations that present different molecular and physiological functions. The ABCC4 High subpopulation demonstrates significant EMT reprogramming, RNA metabolism and high response to DNA damage stimuli. The ABCG2 High subpopulation may resist the anti-EGFR therapy, presenting higher proteolytical activity.
Keywords: ABCC4; ABCG2; CRC; CRC diagnostic and prognostic biomarkers; CRC subpopulations; immune cell infiltration; metastasis.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures




Similar articles
-
Implications of ABCC4-Mediated cAMP Eflux for CRC Migration.Cancers (Basel). 2020 Nov 27;12(12):3547. doi: 10.3390/cancers12123547. Cancers (Basel). 2020. PMID: 33261018 Free PMC article.
-
Abcc4 together with abcb1 and abcg2 form a robust cooperative drug efflux system that restricts the brain entry of camptothecin analogues.Clin Cancer Res. 2013 Apr 15;19(8):2084-95. doi: 10.1158/1078-0432.CCR-12-3105. Epub 2013 Mar 5. Clin Cancer Res. 2013. PMID: 23461902
-
cGMP transport by vesicles from human and mouse erythrocytes.FEBS J. 2007 Jan;274(2):439-50. doi: 10.1111/j.1742-4658.2006.05591.x. FEBS J. 2007. PMID: 17229149
-
Expression of ABCG2 associated with tumor response in metastatic colorectal cancer patients receiving first-line FOLFOX therapy--preliminary evidence.Int J Biol Markers. 2013 Jun 28;28(2):182-6. doi: 10.5301/jbm.5000004. Int J Biol Markers. 2013. PMID: 23558935
-
Implications of ABCG2 Expression on Irinotecan Treatment of Colorectal Cancer Patients: A Review.Int J Mol Sci. 2017 Sep 7;18(9):1926. doi: 10.3390/ijms18091926. Int J Mol Sci. 2017. PMID: 28880238 Free PMC article. Review.
Cited by
-
The Expression Level of SOX Family Transcription Factors' mRNA as a Diagnostic Marker for Osteoarthritis.J Clin Med. 2025 Feb 11;14(4):1176. doi: 10.3390/jcm14041176. J Clin Med. 2025. PMID: 40004707 Free PMC article.
-
A preliminary bioinformatic screen to identify SRI SMC2 PSIP1 TLE4 and MSX1 as potential diagnostic and prognostic markers of osteoarthritis.Sci Rep. 2025 Aug 20;15(1):30583. doi: 10.1038/s41598-025-13575-2. Sci Rep. 2025. PMID: 40835866 Free PMC article.
References
-
- Hsu H.-H., Chen M.-C., Baskaran R., Lin Y.-M., Day C.H., Lin Y.-J., Tu C.-C., Vijaya Padma V., Kuo W.-W., Huang C.-Y. Oxaliplatin resistance in colorectal cancer cells is mediated via activation of ABCG2 to alleviate ER stress induced apoptosis. J. Cell. Physiol. 2018;233:5458–5467. doi: 10.1002/jcp.26406. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

