Comprehensive Profiling and Therapeutic Insights into Differentially Expressed Genes in Hepatocellular Carcinoma
- PMID: 38067357
- PMCID: PMC10705715
- DOI: 10.3390/cancers15235653
Comprehensive Profiling and Therapeutic Insights into Differentially Expressed Genes in Hepatocellular Carcinoma
Abstract
Background: Drug repurposing is a strategy that complements the conventional approach of developing new drugs. Hepatocellular carcinoma (HCC) is a highly prevalent type of liver cancer, necessitating an in-depth understanding of the underlying molecular alterations for improved treatment. Methods: We searched for a vast array of microarray experiments in addition to RNA-seq data. Through rigorous filtering processes, we have identified highly representative differentially expressed genes (DEGs) between tumor and non-tumor liver tissues and identified a distinct class of possible new candidate drugs. Results: Functional enrichment analysis revealed distinct biological processes associated with metal ions, including zinc, cadmium, and copper, potentially implicating chronic metal ion exposure in tumorigenesis. Conversely, up-regulated genes are associated with mitotic events and kinase activities, aligning with the relevance of kinases in HCC. To unravel the regulatory networks governing these DEGs, we employed topological analysis methods, identifying 25 hub genes and their regulatory transcription factors. In the pursuit of potential therapeutic options, we explored drug repurposing strategies based on computational approaches, analyzing their potential to reverse the expression patterns of key genes, including AURKA, CCNB1, CDK1, RRM2, and TOP2A. Potential therapeutic chemicals are alvocidib, AT-7519, kenpaullone, PHA-793887, JNJ-7706621, danusertibe, doxorubicin and analogues, mitoxantrone, podofilox, teniposide, and amonafide. Conclusion: This multi-omic study offers a comprehensive view of DEGs in HCC, shedding light on potential therapeutic targets and drug repurposing opportunities.
Keywords: bioinformatics; drug repurposing; druggable genes; liver cancer; reverse expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
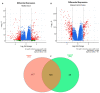



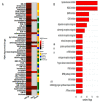


Similar articles
-
Integrated Bioinformatics Analysis for the Screening of Hub Genes and Therapeutic Drugs in Hepatocellular Carcinoma.Curr Pharm Biotechnol. 2023;24(8):1035-1058. doi: 10.2174/1389201023666220628113452. Curr Pharm Biotechnol. 2023. PMID: 35762549
-
A systematic study on the treatment of hepatitis B-related hepatocellular carcinoma with drugs based on bioinformatics and key target reverse network pharmacology and experimental verification.Infect Agent Cancer. 2023 Jul 1;18(1):41. doi: 10.1186/s13027-023-00520-z. Infect Agent Cancer. 2023. PMID: 37393234 Free PMC article.
-
[Bioinformatics analysis of core differentially expressed genes in hepatitis B virus-related hepatocellular carcinoma].Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2022 Nov 21;34(5):507-513. doi: 10.16250/j.32.1374.2021292. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2022. PMID: 36464255 Chinese.
-
Identification of key genes and pathways in hepatocellular carcinoma: A preliminary bioinformatics analysis.Medicine (Baltimore). 2019 Feb;98(5):e14287. doi: 10.1097/MD.0000000000014287. Medicine (Baltimore). 2019. PMID: 30702595 Free PMC article.
-
Screening Hub Genes as Prognostic Biomarkers of Hepatocellular Carcinoma by Bioinformatics Analysis.Cell Transplant. 2019 Dec;28(1_suppl):76S-86S. doi: 10.1177/0963689719893950. Epub 2019 Dec 11. Cell Transplant. 2019. PMID: 31822116 Free PMC article.
Cited by
-
Identifying key genes involved in HBV-related hepatocellular carcinoma: diagnose, prognosis, interaction and immune analysis.Discov Oncol. 2025 Aug 9;16(1):1510. doi: 10.1007/s12672-025-03320-6. Discov Oncol. 2025. PMID: 40782246 Free PMC article.
-
Integrating CT Radiomics and Clinical Features to Optimize TACE Technique Decision-Making in Hepatocellular Carcinoma.Cancers (Basel). 2025 Mar 5;17(5):893. doi: 10.3390/cancers17050893. Cancers (Basel). 2025. PMID: 40075740 Free PMC article.
-
Unveiling the Therapeutic Potential of Targeting RRM2 in Hepatocellular Carcinoma: An Integrated In Silico and In Vitro Study.Funct Integr Genomics. 2025 Jun 10;25(1):123. doi: 10.1007/s10142-025-01630-0. Funct Integr Genomics. 2025. PMID: 40493217 Free PMC article.
-
Integrating TCGA and Single-Cell Sequencing Data for Hepatocellular Carcinoma: A Novel Glycosylation (GLY)/Tumor Microenvironment (TME) Classifier to Predict Prognosis and Immunotherapy Response.Metabolites. 2024 Jan 13;14(1):51. doi: 10.3390/metabo14010051. Metabolites. 2024. PMID: 38248854 Free PMC article.
-
Key oncogenes and candidate drugs for hepatitis-B-driven hepatocellular carcinoma progression.Discov Oncol. 2025 Feb 4;16(1):116. doi: 10.1007/s12672-025-01851-6. Discov Oncol. 2025. PMID: 39903352 Free PMC article.
References
-
- Parra N.S., Ross H.M., Khan A., Wu M., Goldberg R., Shah L., Mukhtar S., Beiriger J., Gerber A., Halegoua-DeMarzio D. Advancements in the Diagnosis of Hepatocellular Carcinoma. Int. J. Transl. Med. 2023;3:51–65. doi: 10.3390/ijtm3010005. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

