Schlafen Family Intra-Regulation by IFN-α2 in Triple-Negative Breast Cancer
- PMID: 38067362
- PMCID: PMC10705374
- DOI: 10.3390/cancers15235658
Schlafen Family Intra-Regulation by IFN-α2 in Triple-Negative Breast Cancer
Abstract
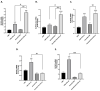
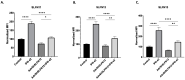
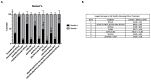
Triple-negative breast cancer (TNBC) has a poor prognosis and no targeted therapy for treatment. The Schlafen gene family, particularly SLFN12, critically mediates TNBC biology. Higher expression of SLFN12 correlates with decreased TNBC viability and increased chemosensitivity and patient survival, yet no treatment is known to upregulate SLFN12 in TNBC. We hypothesized that Interferon-α (IFN-α2) upregulates SLFN12 in TNBC, subsequently reducing cell viability. We utilized short hairpin adenovirus to knockout SLFN12 (AdvShSLFN12) in MDA-MB-231, Hs-578T, and BT-549 TNBC cells. Cells were treated with AdvShSLFN12 and IFN-α2. After treatment, TNBC cell viability, SLFN family mRNA, and protein expression were analyzed. Treating TNBC cells with IFN-α2 increased SLFN12 expression and reduced cell viability. However, when AdvShSLFN12 knocked down SLFN12 during IFN-α2 treatment, TNBC cell viability was still reduced. We, therefore, investigated the potential involvement of other SLFN members IFN-α2 effects on cell viability. IFN-α2 increased SLFN5, SLFN12-Like, and SLFN14 but not SLFN11 or SLFN13. During AdvShSLFN12 + IFN-α2 treatment, the expressions of SLFN5, SLFN12-Like, and SLFN14 further increased. However, when siRNA knocked down SLFN5, SLFN12-Like, and SLFN14, the IFN-α2 reduction in viability was blunted. Although the interpretation of these results may be limited by the potential interactions between different siRNAs, these data suggest a complex regulatory signaling cascade among SLFN family members. Targeting this cascade to manipulate SLFN levels may, in the future, offer the potential to manipulate the chemosensitivity of TNBC tumors.
Keywords: AdvShSLFN12; IFN-?2; Schlafen 12; TNBC; cell viability.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of this study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
SLFN12 Expression Significantly Effects the Response to Chemotherapy Drugs in Triple-Negative Breast Cancer.Cancers (Basel). 2024 Nov 16;16(22):3848. doi: 10.3390/cancers16223848. Cancers (Basel). 2024. PMID: 39594803 Free PMC article.
-
Schlafen family is a prognostic biomarker and corresponds with immune infiltration in gastric cancer.Front Immunol. 2022 Aug 25;13:922138. doi: 10.3389/fimmu.2022.922138. eCollection 2022. Front Immunol. 2022. PMID: 36090985 Free PMC article.
-
Expression and regulation of Schlafen (SLFN) family members in primary human monocytes, monocyte-derived dendritic cells and T cells.Results Immunol. 2015 Oct 22;5:23-32. doi: 10.1016/j.rinim.2015.10.001. eCollection 2015. Results Immunol. 2015. PMID: 26623250 Free PMC article.
-
Structural, molecular, and functional insights into Schlafen proteins.Exp Mol Med. 2022 Jun;54(6):730-738. doi: 10.1038/s12276-022-00794-0. Epub 2022 Jun 29. Exp Mol Med. 2022. PMID: 35768579 Free PMC article. Review.
-
Overview of Structural and Functional Insights of SLFN12 Associated With Different Diseases.Cureus. 2024 May 2;16(5):e59515. doi: 10.7759/cureus.59515. eCollection 2024 May. Cureus. 2024. PMID: 38832156 Free PMC article. Review.
Cited by
-
Research progress of the SLFN family in malignant tumors.Front Oncol. 2024 Nov 4;14:1468484. doi: 10.3389/fonc.2024.1468484. eCollection 2024. Front Oncol. 2024. PMID: 39558948 Free PMC article. Review.
-
Schlafens: Emerging Therapeutic Targets.Cancers (Basel). 2024 May 9;16(10):1805. doi: 10.3390/cancers16101805. Cancers (Basel). 2024. PMID: 38791884 Free PMC article. Review.
References
-
- American Cancer Society . About Breast Cancer. American Cancer Society; New York, NY, USA: 2023.
-
- Baranova A., Krasnoselskyi M., Starikov V., Kartashov S., Zhulkevych I., Vlasenko V., Oleshko K., Bilodid O., Sadchikova M., Vinnyk Y. Triple-Negative Breast Cancer: Current Treatment Strategies and Factors of Negative Prognosis. J. Med. Life. 2022;2022:153–161. doi: 10.25122/jml-2021-0108. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

