Influence of Amino Acid Substitutions in ApoMb on Different Stages of Unfolding of Amyloids
- PMID: 38067466
- PMCID: PMC10707739
- DOI: 10.3390/molecules28237736
Influence of Amino Acid Substitutions in ApoMb on Different Stages of Unfolding of Amyloids
Abstract
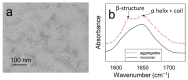
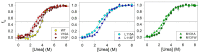
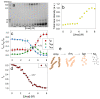
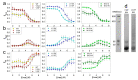
To date, most research on amyloid aggregation has focused on describing the structure of amyloids and the kinetics of their formation, while the conformational stability of fibrils remains insufficiently explored. The aim of this work was to investigate the effect of amino acid substitutions on the stability of apomyoglobin (ApoMb) amyloids. A study of the amyloid unfolding of ApoMb and its six mutant variants by urea has been carried out. Changes in the structural features of aggregates during unfolding were recorded by far-UV CD and native electrophoresis. It was shown that during the initial stage of denaturation, amyloids' secondary structure partially unfolds. Then, the fibrils undergo dissociation and form intermediate aggregates weighing approximately 1 MDa, which at the last stage of unfolding decompose into 18 kDa monomeric unfolded molecules. The results of unfolding transitions suggest that the stability of the studied amyloids relative to the intermediate aggregates and of the latter relative to unfolded monomers is higher for ApoMb variants with substitutions that increase the hydrophobicity of the residues. The results presented provide a new insight into the mechanism of stabilization of protein aggregates and can serve as a base for further investigations of the amyloids' stability.
Keywords: amyloid stability; apomyoglobin; hydrophobicity; native electrophoresis; unfolding transition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
sw ApoMb Amyloid Aggregation under Nondenaturing Conditions: The Role of Native Structure Stability.Biophys J. 2017 Sep 5;113(5):991-1001. doi: 10.1016/j.bpj.2017.07.011. Biophys J. 2017. PMID: 28877500 Free PMC article.
-
[Amyloid Core Wild-Type Apomyoglobin and Its Mutant Variants Is Formed by Different Regions of the Polypeptide Chain].Mol Biol (Mosk). 2018 Jan-Feb;52(1):51-61. doi: 10.7868/S0026898418010081. Mol Biol (Mosk). 2018. PMID: 29512636 Russian.
-
[Equilibrium unfolding of mutant apomyoglobins with substitutions of conserved nonfunctional residues by alanine].Mol Biol (Mosk). 2007 Jul-Aug;41(4):674-80. doi: 10.1134/s0026893307040231. Mol Biol (Mosk). 2007. PMID: 17936988 Russian.
-
The folding process of apomyoglobin.Protein Pept Lett. 2005 Apr;12(3):229-34. doi: 10.2174/0929866053587174. Protein Pept Lett. 2005. PMID: 15777270 Review.
-
Amyloid fibril formation and disaggregation of fragment 1-29 of apomyoglobin: insights into the effect of pH on protein fibrillogenesis.J Mol Biol. 2007 Apr 13;367(5):1237-45. doi: 10.1016/j.jmb.2007.01.072. Epub 2007 Feb 3. J Mol Biol. 2007. PMID: 17320902 Review.
Cited by
-
Precise modulation of protein refolding by rationally designed covalent organic frameworks.Nat Commun. 2025 May 3;16(1):4122. doi: 10.1038/s41467-025-59368-z. Nat Commun. 2025. PMID: 40316523 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

