Determination and Kinetic Characterization of a New Potential Inhibitor for AmsI Protein Tyrosine Phosphatase from the Apple Pathogen Erwinia amylovora
- PMID: 38067503
- PMCID: PMC10708540
- DOI: 10.3390/molecules28237774
Determination and Kinetic Characterization of a New Potential Inhibitor for AmsI Protein Tyrosine Phosphatase from the Apple Pathogen Erwinia amylovora
Abstract
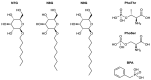
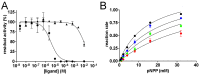
Erwinia amylovora is a Gram-negative bacterium, responsible for the fire blight disease in Rosaceae plants. Its virulence is correlated with the production of an exopolysaccharide (EPS) called amylovoran, which protects the bacterium from the surrounding environment and helps its diffusion inside the host. Amylovoran biosynthesis relies on the expression of twelve genes clustered in the ams operon. One of these genes, amsI, encodes for a Low Molecular Weight Protein Tyrosine Phosphatase (LMW-PTP) called EaAmsI, which plays a key role in the regulation of the EPS production pathway. For this reason, EaAmsI was chosen in this work as a target for the development of new antibacterial agents against E. amylovora. To achieve this aim, a set of programs (DOCK6, OpenEye FRED) was selected to perform a virtual screening using a database of ca. 700 molecules. The six best-scoring compounds identified were tested in in vitro assays. A complete inhibition kinetic characterization carried out on the most promising molecule (n-Heptyl β-D-glucopyranoside, N7G) showed an inhibition constant of 7.8 ± 0.6 µM. This study represents an initial step towards the development of new EaAmsI inhibitors able to act as antibacterial agents against E. amylovora infections.
Keywords: EPS production regulation; Erwinia amylovora; amylovoran; exopolysaccharide; fire blight; in vitro assays; inhibition constant; molecular docking; protein tyrosine phosphatase; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
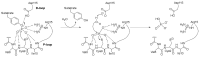

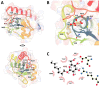
Similar articles
-
Virulence Genetics of an Erwinia amylovora Putative Polysaccharide Transporter Family Member.J Bacteriol. 2020 Oct 22;202(22):e00390-20. doi: 10.1128/JB.00390-20. Print 2020 Oct 22. J Bacteriol. 2020. PMID: 32839177 Free PMC article.
-
Cloning, purification, crystallization and 1.57 Å resolution X-ray data analysis of AmsI, the tyrosine phosphatase controlling amylovoran biosynthesis in the plant pathogen Erwinia amylovora.Acta Crystallogr F Struct Biol Commun. 2014 Dec 1;70(Pt 12):1693-6. doi: 10.1107/S2053230X14024947. Epub 2014 Nov 28. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25484228 Free PMC article.
-
Characterization and 1.57 Å resolution structure of the key fire blight phosphatase AmsI from Erwinia amylovora.Acta Crystallogr F Struct Biol Commun. 2016 Dec 1;72(Pt 12):903-910. doi: 10.1107/S2053230X16018781. Epub 2016 Nov 30. Acta Crystallogr F Struct Biol Commun. 2016. PMID: 27917839 Free PMC article.
-
Molecular genetics of Erwinia amylovora involved in the development of fire blight.FEMS Microbiol Lett. 2005 Dec 15;253(2):185-92. doi: 10.1016/j.femsle.2005.09.051. Epub 2005 Oct 13. FEMS Microbiol Lett. 2005. PMID: 16253442 Review.
-
Virulence Factors of Erwinia amylovora: A Review.Int J Mol Sci. 2015 Jun 5;16(6):12836-54. doi: 10.3390/ijms160612836. Int J Mol Sci. 2015. PMID: 26057748 Free PMC article. Review.
References
-
- Vanneste J.L. Fire Blight: The Disease and Its Causative Agent, Erwinia Amylovora. CABI; Wallingford, UK: 2000.
-
- Zeng Q., Cui Z., Wang J., Childs K.L., Sundin G.W., Cooley D.R., Yang C., Garofalo E., Eaton A., Huntley R.B., et al. Comparative Genomics of Spiraeoideae-infecting Erwinia amylovora Strains Provides Novel Insight to Genetic Diversity and Identifies the Genetic Basis of a Low-virulence Strain. Mol. Plant Pathol. 2018;19:1652–1666. doi: 10.1111/mpp.12647. - DOI - PMC - PubMed
-
- Myung I.-S., Lee J.-Y., Yun M.-J., Lee Y.-H., Lee Y.-K., Park D.-H., Oh C.-S. Fire Blight of Apple, Caused by Erwinia Amylovora, a New Disease in Korea. Plant Dis. 2016;100:1774. doi: 10.1094/PDIS-01-16-0024-PDN. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

