Mmp12 Is Translationally Regulated in Macrophages during the Course of Inflammation
- PMID: 38069304
- PMCID: PMC10707645
- DOI: 10.3390/ijms242316981
Mmp12 Is Translationally Regulated in Macrophages during the Course of Inflammation
Abstract
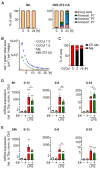
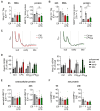
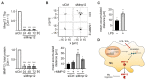
Despite the importance of rapid adaptive responses in the course of inflammation and the notion that post-transcriptional regulation plays an important role herein, relevant translational alterations, especially during the resolution phase, remain largely elusive. In the present study, we analyzed translational changes in inflammatory bone marrow-derived macrophages upon resolution-promoting efferocytosis. Total RNA-sequencing confirmed that apoptotic cell phagocytosis induced a pro-resolution signature in LPS/IFNγ-stimulated macrophages (Mϕ). While inflammation-dependent transcriptional changes were relatively small between efferocytic and non-efferocytic Mϕ; considerable differences were observed at the level of de novo synthesized proteins. Interestingly, translationally regulated targets in response to inflammatory stimuli were mostly downregulated, with only minimal impact of efferocytosis. Amongst these targets, pro-resolving matrix metallopeptidase 12 (Mmp12) was identified as a translationally repressed candidate during early inflammation that recovered during the resolution phase. Functionally, reduced MMP12 production enhanced matrix-dependent migration of Mϕ. Conclusively, translational control of MMP12 emerged as an efficient strategy to alter the migratory properties of Mϕ throughout the inflammatory response, enabling Mϕ migration within the early inflammatory phase while restricting migration during the resolution phase.
Keywords: Mmp12; efferocytosis; inflammation; macrophage; resolution; translation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Savill J.S., Wyllie A.H., Henson J.E., Walport M.J., Henson P.M., Haslett C. Macrophage phagocytosis of aging neutrophils in inflammation. Programmed cell death in the neutrophil leads to its recognition by macrophages. J. Clin. Investig. 1989;83:865–875. doi: 10.1172/JCI113970. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

