Genome-Wide Identification and Comprehensive Analysis of the FtsH Gene Family in Soybean (Glycine max)
- PMID: 38069319
- PMCID: PMC10707429
- DOI: 10.3390/ijms242316996
Genome-Wide Identification and Comprehensive Analysis of the FtsH Gene Family in Soybean (Glycine max)
Abstract
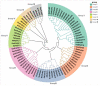
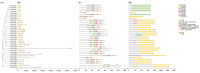
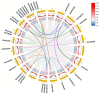
The filamentation temperature-sensitive H (FtsH) gene family is critical in regulating plant chloroplast development and photosynthesis. It plays a vital role in plant growth, development, and stress response. Although FtsH genes have been identified in a wide range of plants, there is no detailed study of the FtsH gene family in soybean (Glycine max). Here, we identified 34 GmFtsH genes, which could be categorized into eight groups, and GmFtsH genes in the same group had similar structures and conserved protein motifs. We also performed intraspecific and interspecific collinearity analysis and found that the GmFtsH family has large-scale gene duplication and is more closely related to Arabidopsis thaliana. Cis-acting elements analysis in the promoter region of the GmFtsH genes revealed that most genes contain developmental and stress response elements. Expression patterns based on transcriptome data and real-time reverse transcription quantitative PCR (qRT-PCR) showed that most of the GmFtsH genes were expressed at the highest levels in leaves. Then, GO enrichment analysis indicated that GmFtsH genes might function as a protein hydrolase. In addition, the GmFtsH13 protein was confirmed to be localized in chloroplasts by a transient expression experiment in tobacco. Taken together, the results of this study lay the foundation for the functional determination of GmFtsH genes and help researchers further understand the regulatory network in soybean leaf development.
Keywords: FtsH family; chloroplast; protein hydrolysis enzyme; soybean.
Conflict of interest statement
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Figures






Similar articles
-
Genome-wide identification and comprehensive analysis of the FtsH gene family in wheat.Mol Biol Rep. 2025 Feb 3;52(1):186. doi: 10.1007/s11033-025-10243-6. Mol Biol Rep. 2025. PMID: 39899074
-
Genome-wide analysis of filamentous temperature-sensitive H protease (ftsH) gene family in soybean.BMC Genomics. 2024 May 27;25(1):524. doi: 10.1186/s12864-024-10389-w. BMC Genomics. 2024. PMID: 38802777 Free PMC article.
-
Genome-wide identification and expression analysis of the ftsH protein family and its response to abiotic stress in Nicotiana tabacum L.BMC Genomics. 2022 Jul 12;23(1):503. doi: 10.1186/s12864-022-08719-x. BMC Genomics. 2022. PMID: 35831784 Free PMC article.
-
Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis.BMC Genomics. 2015 Jul 11;16(1):520. doi: 10.1186/s12864-015-1730-y. BMC Genomics. 2015. PMID: 26162601 Free PMC article.
-
Genome-wide identification, evolution, and expression analysis of carbonic anhydrases genes in soybean (Glycine max).Funct Integr Genomics. 2023 Jan 14;23(1):37. doi: 10.1007/s10142-023-00966-9. Funct Integr Genomics. 2023. PMID: 36639600 Review.
Cited by
-
Genome-wide identification and comprehensive analysis of the FtsH gene family in wheat.Mol Biol Rep. 2025 Feb 3;52(1):186. doi: 10.1007/s11033-025-10243-6. Mol Biol Rep. 2025. PMID: 39899074
-
A mitochondrial metalloprotease FtsH4 is required for symbiotic nitrogen fixation in Lotus japonicus nodules.Sci Rep. 2024 Nov 11;14(1):27578. doi: 10.1038/s41598-024-78295-5. Sci Rep. 2024. PMID: 39528551 Free PMC article.
-
Genome-wide identification of the ATP-dependent zinc metalloprotease (FtsH) in Triticeae species reveals that TaFtsH-1 regulates cadmium tolerance in Triticum aestivum.PLoS One. 2024 Dec 31;19(12):e0316486. doi: 10.1371/journal.pone.0316486. eCollection 2024. PLoS One. 2024. PMID: 39739686 Free PMC article.
-
The members of zinc finger-homeodomain (ZF-HD) transcription factors are associated with abiotic stresses in soybean: insights from genomics and expression analysis.BMC Plant Biol. 2025 Jan 14;25(1):56. doi: 10.1186/s12870-024-06028-x. BMC Plant Biol. 2025. PMID: 39810081 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

