Genome-Wide Identification and Comprehensive Analysis of the GASA Gene Family in Peanuts (Arachis hypogaea L.) under Abiotic Stress
- PMID: 38069439
- PMCID: PMC10707693
- DOI: 10.3390/ijms242317117
Genome-Wide Identification and Comprehensive Analysis of the GASA Gene Family in Peanuts (Arachis hypogaea L.) under Abiotic Stress
Abstract
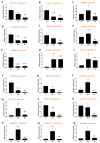
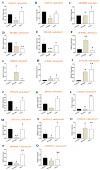
Peanut (Arachis hypogaea L.) is a globally cultivated crop of significant economic and nutritional importance. The role of gibberellic-acid-stimulated Arabidopsis (GASA) family genes is well established in plant growth, development, and biotic and abiotic stress responses. However, there is a gap in understanding the function of GASA proteins in cultivated peanuts, particularly in response to abiotic stresses such as drought and salinity. Thus, we conducted comprehensive in silico analyses to identify and verify the existence of 40 GASA genes (termed AhGASA) in cultivated peanuts. Subsequently, we conducted biological experiments and performed expression analyses of selected AhGASA genes to elucidate their potential regulatory roles in response to drought and salinity. Phylogenetic analysis revealed that AhGASA genes could be categorized into four distinct subfamilies. Under normal growth conditions, selected AhGASA genes exhibited varying expressions in young peanut seedling leaves, stems, and roots tissues. Notably, our findings indicate that certain AhGASA genes were downregulated under drought stress but upregulated under salt stress. These results suggest that specific AhGASA genes are involved in the regulation of salt or drought stress. Further functional characterization of the upregulated genes under both drought and salt stress will be essential to confirm their regulatory roles in this context. Overall, our findings provide compelling evidence of the involvement of AhGASA genes in the mechanisms of stress tolerance in cultivated peanuts. This study enhances our understanding of the functions of AhGASA genes in response to abiotic stress and lays the groundwork for future investigations into the molecular characterization of AhGASA genes.
Keywords: GASA; drought; expression analysis; gibberellin; peanut; phylogenetic analysis; salt stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Rezaee S., Ahmadizadeh M., Heidari P. Genome-wide characterization, expression profiling, and post-transcriptional study of GASA gene family. Gene Rep. 2020;20:100795. doi: 10.1016/j.genrep.2020.100795. - DOI
-
- Muhammad I., Li W.-Q., Jing X.-Q., Zhou M.-R., Shalmani A., Ali M., Wei X.-Y., Sharif R., Liu W.-T., Chen K.-M. A systematic in silico prediction of gibberellic acid stimulated GASA family members: A novel small peptide contributes to floral architecture and transcriptomic changes induced by external stimuli in rice. J. Plant Physiol. 2019;234–235:117–132. doi: 10.1016/j.jplph.2019.02.005. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

