The long non-coding RNA HOTAIR contributes to joint-specific gene expression in rheumatoid arthritis
- PMID: 38071204
- PMCID: PMC10710443
- DOI: 10.1038/s41467-023-44053-w
The long non-coding RNA HOTAIR contributes to joint-specific gene expression in rheumatoid arthritis
Abstract
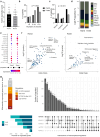
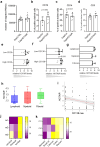
Although patients with rheumatoid arthritis (RA) typically exhibit symmetrical joint involvement, some patients develop alternative disease patterns in response to treatment, suggesting that different molecular mechanism may underlie disease progression depending on joint location. Here, we identify joint-specific changes in RA synovium and synovial fibroblasts (SF) between knee and hand joints. We show that the long non-coding RNA HOTAIR, which is only expressed in knee SF, regulates more than 50% of this site-specific gene expression in SF. HOTAIR is downregulated after stimulation with pro-inflammatory cytokines and is expressed at lower levels in knee samples from patients with RA, compared with osteoarthritis. Knockdown of HOTAIR in knee SF increases PI-Akt signalling and IL-6 production, but reduces Wnt signalling. Silencing HOTAIR inhibits the migratory function of SF, decreases SF-mediated osteoclastogenesis, and increases the recruitment of B cells by SF. We propose that HOTAIR is an important epigenetic factor in joint-specific gene expression in RA.
© 2023. The Author(s).
Conflict of interest statement
M.E., R.M., L.K., A.K., M.T., T.R., C.G., M.H., M.M., L.M., C.P., K.B., A.L., G.K., K.K., P.S., M.F.B., S.G.E., M.S., G.K., M.A., and C.O. declare no competing interests. O.D. has/had relationships with the following companies in the area of potential treatments for systemic sclerosis and its complications in the last three calendar years: Speaker fee: Bayer, Boehringer Ingelheim, Janssen, Medscape Consultancy fee: 4P-Pharma, Abbvie, Acceleron, Alcimed, Altavant Siences, Amgen, AnaMar, Arxx, AstraZeneca, Baecon, Blade, Bayer, Boehringer Ingelheim, Corbus, CSL Behring, Galapagos, Glenmark, Horizon, Inventiva, Kymera, Lupin, Miltenyi Biotec, Mitsubishi Tanabe, MSD, Novartis, Prometheus, Redxpharna, Roivant, Sanofi and Topadur Research Grants: Kymera, Mitsubishi Tanabe, Boehringer Ingelheim Oliver Distler has/had relationships with the following companies in the area of potential treatments for dermatomyositis and its complications in the last three calendar years: Consultancy fee for rheumatology topic: Pfizer (2021) Oliver Distler has/had relationships with the following companies in the area of potential treatments for arthritides in the last three Consultancy fee: Abbvie.
Figures







References
-
- Bijlsma, J. W. J. et al. EULAR textbook on rheumatic diseases. BMJ Publishing Group United Kingdom (2018).
-
- van der Woude D. van der Helm-van Mil AHM. Update on the epidemiology, risk factors, and disease outcomes of rheumatoid arthritis. Best practice & research. Clin. Rheumatol. 2018;32:174–187. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

