RAAS-deficient organoids indicate delayed angiogenesis as a possible cause for autosomal recessive renal tubular dysgenesis
- PMID: 38071212
- PMCID: PMC10710424
- DOI: 10.1038/s41467-023-43795-x
RAAS-deficient organoids indicate delayed angiogenesis as a possible cause for autosomal recessive renal tubular dysgenesis
Abstract
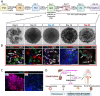
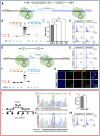
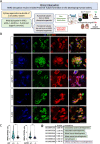
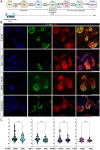
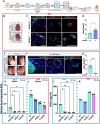
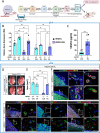
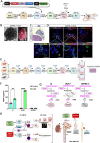
Autosomal Recessive Renal Tubular Dysgenesis (AR-RTD) is a fatal genetic disorder characterized by complete absence or severe depletion of proximal tubules (PT) in patients harboring pathogenic variants in genes involved in the Renin-Angiotensin-Aldosterone System. To uncover the pathomechanism of AR-RTD, differentiation of ACE-/- and AGTR1-/- induced pluripotent stem cells (iPSCs) and AR-RTD patient-derived iPSCs into kidney organoids is leveraged. Comprehensive marker analyses show that both mutant and control organoids generate indistinguishable PT in vitro under normoxic (21% O2) or hypoxic (2% O2) conditions. Fully differentiated (d24) AGTR1-/- and control organoids transplanted under the kidney capsule of immunodeficient mice engraft and mature well, as do renal vesicle stage (d14) control organoids. By contrast, d14 AGTR1-/- organoids fail to engraft due to insufficient pro-angiogenic VEGF-A expression. Notably, growth under hypoxic conditions induces VEGF-A expression and rescues engraftment of AGTR1-/- organoids at d14, as does ectopic expression of VEGF-A. We propose that PT dysgenesis in AR-RTD is primarily a non-autonomous consequence of delayed angiogenesis, starving PT at a critical time in their development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

