Generating high-quality plant and fish reference genomes from field-collected specimens by optimizing preservation
- PMID: 38071270
- PMCID: PMC10710401
- DOI: 10.1038/s42003-023-05615-2
Generating high-quality plant and fish reference genomes from field-collected specimens by optimizing preservation
Abstract
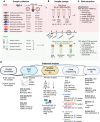
Sample preservation often impedes efforts to generate high-quality reference genomes or pangenomes for Earth's more than 2 million plant and animal species due to nucleotide degradation. Here we compare the impacts of storage methods including solution type, temperature, and time on DNA quality and Oxford Nanopore long-read sequencing quality in 9 fish and 4 plant species. We show 95% ethanol largely protects against degradation for fish blood (22 °C, ≤6 weeks) and plant tissue (4 °C, ≤3 weeks). From this furthest storage timepoint, we assemble high-quality reference genomes of 3 fish and 2 plant species with contiguity (contig N50) and completeness (BUSCO) that achieve the Vertebrate Genome Project benchmarking standards. For epigenetic applications, we also report methylation frequency compared to liquid nitrogen control. The results presented here remove the necessity for cryogenic storage in many long read applications and provide a framework for future studies focused on sampling in remote locations, which may represent a large portion of the future sequencing of novel organisms.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Method of the Year 2022: long-read sequencing. Nat. Methods20, 1 (2023). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

