Loss of cohesin regulator PDS5A reveals repressive role of Polycomb loops
- PMID: 38071364
- PMCID: PMC10710464
- DOI: 10.1038/s41467-023-43869-w
Loss of cohesin regulator PDS5A reveals repressive role of Polycomb loops
Abstract
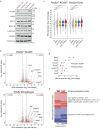
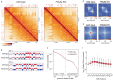
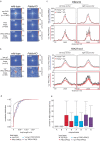
Polycomb Repressive Complexes 1 and 2 (PRC1, PRC2) are conserved epigenetic regulators that promote transcriptional gene silencing. PRC1 and PRC2 converge on shared targets, catalyzing repressive histone modifications. Additionally, a subset of PRC1/PRC2 targets engage in long-range interactions whose functions in gene silencing are poorly understood. Using a CRISPR screen in mouse embryonic stem cells, we found that the cohesin regulator PDS5A links transcriptional silencing by Polycomb and 3D genome organization. PDS5A deletion impairs cohesin unloading and results in derepression of a subset of endogenous PRC1/PRC2 target genes. Importantly, derepression is not linked to loss of Polycomb chromatin domains. Instead, PDS5A removal causes aberrant cohesin activity leading to ectopic insulation sites, which disrupt the formation of ultra-long Polycomb loops. We show that these loops are important for robust silencing at a subset of PRC1/PRC2 target genes and that maintenance of cohesin-dependent genome architecture is critical for Polycomb regulation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

