Interrogation and validation of the interactome of neuronal Munc18-interacting Mint proteins with AlphaFold2
- PMID: 38072052
- PMCID: PMC10820826
- DOI: 10.1016/j.jbc.2023.105541
Interrogation and validation of the interactome of neuronal Munc18-interacting Mint proteins with AlphaFold2
Abstract
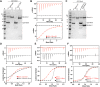
Munc18-interacting proteins (Mints) are multidomain adaptors that regulate neuronal membrane trafficking, signaling, and neurotransmission. Mint1 and Mint2 are highly expressed in the brain with overlapping roles in the regulation of synaptic vesicle fusion required for neurotransmitter release by interacting with the essential synaptic protein Munc18-1. Here, we have used AlphaFold2 to identify and then validate the mechanisms that underpin both the specific interactions of neuronal Mint proteins with Munc18-1 as well as their wider interactome. We found that a short acidic α-helical motif within Mint1 and Mint2 is necessary and sufficient for specific binding to Munc18-1 and binds a conserved surface on Munc18-1 domain3b. In Munc18-1/2 double knockout neurosecretory cells, mutation of the Mint-binding site reduces the ability of Munc18-1 to rescue exocytosis, and although Munc18-1 can interact with Mint and Sx1a (Syntaxin1a) proteins simultaneously in vitro, we find that they have mutually reduced affinities, suggesting an allosteric coupling between the proteins. Using AlphaFold2 to then examine the entire cellular network of putative Mint interactors provides a structural model for their assembly with a variety of known and novel regulatory and cargo proteins including ADP-ribosylation factor (ARF3/ARF4) small GTPases and the AP3 clathrin adaptor complex. Validation of Mint1 interaction with a new predicted binder TJAP1 (tight junction-associated protein 1) provides experimental support that AlphaFold2 can correctly predict interactions across such large-scale datasets. Overall, our data provide insights into the diversity of interactions mediated by the Mint family and show that Mints may help facilitate a key trigger point in SNARE (soluble N-ethylmaleimide-sensitive factor attachment receptor) complex assembly and vesicle fusion.
Keywords: AP3; APP; ARF3; AlphaFold; CASK; IQSEC1; LRP; Mint; Munc18; SNARE; STXBP1; TJAP1; X11; calsyntenin; neurexin.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures







Similar articles
-
Mints as adaptors. Direct binding to neurexins and recruitment of munc18.J Biol Chem. 2000 Dec 22;275(51):39803-6. doi: 10.1074/jbc.C000656200. J Biol Chem. 2000. PMID: 11036064
-
Mints, Munc18-interacting proteins in synaptic vesicle exocytosis.J Biol Chem. 1997 Dec 12;272(50):31459-64. doi: 10.1074/jbc.272.50.31459. J Biol Chem. 1997. PMID: 9395480
-
Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus.J Neurosci. 2007 Nov 7;27(45):12147-55. doi: 10.1523/JNEUROSCI.3655-07.2007. J Neurosci. 2007. PMID: 17989281 Free PMC article.
-
Neuronal SNARE complex assembly guided by Munc18-1 and Munc13-1.FEBS Open Bio. 2022 Nov;12(11):1939-1957. doi: 10.1002/2211-5463.13394. Epub 2022 Mar 22. FEBS Open Bio. 2022. PMID: 35278279 Free PMC article. Review.
-
Munc18-1 as a key regulator of neurosecretion.J Neurochem. 2010 Oct;115(1):1-10. doi: 10.1111/j.1471-4159.2010.06900.x. J Neurochem. 2010. PMID: 20681955 Review.
Cited by
-
Surface avidity of anionic polypeptide coatings target nanoparticles to cancer-associated amino acid transporters.bioRxiv [Preprint]. 2025 Jul 31:2025.07.28.667320. doi: 10.1101/2025.07.28.667320. bioRxiv. 2025. PMID: 40766676 Free PMC article. Preprint.
-
PPIscreenML: Structure-based screening for protein-protein interactions using AlphaFold.bioRxiv [Preprint]. 2024 Apr 30:2024.03.16.585347. doi: 10.1101/2024.03.16.585347. bioRxiv. 2024. PMID: 38559274 Free PMC article. Preprint.
-
Selecting genes for analysis using historically contingent progress: from RNA changes to protein-protein interactions.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1246. doi: 10.1093/nar/gkae1246. Nucleic Acids Res. 2025. PMID: 39788543 Free PMC article.
-
CRISPR/Cas9: a cutting-edge solution for combatting the fall armyworm, Spodoptera frugiperda.Mol Biol Rep. 2023 Dec 12;51(1):13. doi: 10.1007/s11033-023-08986-1. Mol Biol Rep. 2023. PMID: 38085335 Review.
-
Selecting genes for analysis using historically contingent progress: from RNA changes to protein-protein interactions.bioRxiv [Preprint]. 2024 Oct 19:2024.05.01.592119. doi: 10.1101/2024.05.01.592119. bioRxiv. 2024. Update in: Nucleic Acids Res. 2025 Jan 7;53(1):gkae1246. doi: 10.1093/nar/gkae1246. PMID: 38746289 Free PMC article. Updated. Preprint.
References
-
- Rogelj B., Mitchell J.C., Miller C.C., McLoughlin D.M. The X11/Mint family of adaptor proteins. Brain Res. Rev. 2006;52:305–315. - PubMed
-
- Ho C.S., Marinescu V., Steinhilb M.L., Gaut J.R., Turner R.S., Stuenkel E.L. Synergistic effects of Munc18a and X11 proteins on amyloid precursor protein metabolism. J. Biol. Chem. 2002;277:27021–27028. - PubMed
-
- Lee J.H., Lau K.F., Perkinton M.S., Standen C.L., Shemilt S.J., Mercken L., et al. The neuronal adaptor protein X11alpha reduces Abeta levels in the brains of Alzheimer's APPswe Tg2576 transgenic mice. J. Biol. Chem. 2003;278:47025–47029. - PubMed
-
- Motodate R., Saito Y., Hata S., Suzuki T. Expression and localization of X11 family proteins in neurons. Brain Res. 2016;1646:227–234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

