Highly Accurate Estimation of Cell Type Abundance in Bulk Tissues Based on Single-Cell Reference and Domain Adaptive Matching
- PMID: 38072669
- PMCID: PMC10870031
- DOI: 10.1002/advs.202306329
Highly Accurate Estimation of Cell Type Abundance in Bulk Tissues Based on Single-Cell Reference and Domain Adaptive Matching
Abstract
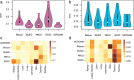
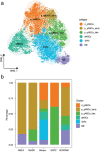
Accurately identifies the cellular composition of complex tissues, which is critical for understanding disease pathogenesis, early diagnosis, and prevention. However, current methods for deconvoluting bulk RNA sequencing (RNA-seq) typically rely on matched single-cell RNA sequencing (scRNA-seq) as a reference, which can be limiting due to differences in sequencing distribution and the potential for invalid information from single-cell references. Hence, a novel computational method named SCROAM is introduced to address these challenges. SCROAM transforms scRNA-seq and bulk RNA-seq into a shared feature space, effectively eliminating distributional differences in the latent space. Subsequently, cell-type-specific expression matrices are generated from the scRNA-seq data, facilitating the precise identification of cell types within bulk tissues. The performance of SCROAM is assessed through benchmarking against simulated and real datasets, demonstrating its accuracy and robustness. To further validate SCROAM's performance, single-cell and bulk RNA-seq experiments are conducted on mouse spinal cord tissue, with SCROAM applied to identify cell types in bulk tissue. Results indicate that SCROAM is a highly effective tool for identifying similar cell types. An integrated analysis of liver cancer and primary glioblastoma is then performed. Overall, this research offers a novel perspective for delivering precise insights into disease pathogenesis and potential therapeutic strategies.
Keywords: deconvolution; tissue heterogeneity; transcriptomics; transfer learning.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Cell-type deconvolution for bulk RNA-seq data using single-cell reference: a comparative analysis and recommendation guideline.Brief Bioinform. 2024 Nov 22;26(1):bbaf031. doi: 10.1093/bib/bbaf031. Brief Bioinform. 2024. PMID: 39899596 Free PMC article.
-
Approximate estimation of cell-type resolution transcriptome in bulk tissue through matrix completion.Brief Bioinform. 2023 Sep 20;24(5):bbad273. doi: 10.1093/bib/bbad273. Brief Bioinform. 2023. PMID: 37529921
-
scAnno: a deconvolution strategy-based automatic cell type annotation tool for single-cell RNA-sequencing data sets.Brief Bioinform. 2023 May 19;24(3):bbad179. doi: 10.1093/bib/bbad179. Brief Bioinform. 2023. PMID: 37183449
-
Fourteen years of cellular deconvolution: methodology, applications, technical evaluation and outstanding challenges.Nucleic Acids Res. 2024 May 22;52(9):4761-4783. doi: 10.1093/nar/gkae267. Nucleic Acids Res. 2024. PMID: 38619038 Free PMC article. Review.
-
Advance and Application of Single-cell Transcriptomics in Auditory Research.Neurosci Bull. 2024 Jul;40(7):963-980. doi: 10.1007/s12264-023-01149-z. Epub 2023 Nov 28. Neurosci Bull. 2024. PMID: 38015350 Free PMC article. Review.
Cited by
-
FusionEncoder: identification of intrinsically disordered regions based on multi-feature fusion.Bioinformatics. 2025 Jul 1;41(7):btaf362. doi: 10.1093/bioinformatics/btaf362. Bioinformatics. 2025. PMID: 40577786 Free PMC article.
-
Identification of DNA N6-methyladenine modifications in the rice genome with a fine-tuned large language model.Front Plant Sci. 2025 Jun 25;16:1626539. doi: 10.3389/fpls.2025.1626539. eCollection 2025. Front Plant Sci. 2025. PMID: 40636005 Free PMC article.
-
NeXtMD: a new generation of machine learning and deep learning stacked hybrid framework for accurate identification of anti-inflammatory peptides.BMC Biol. 2025 Jul 15;23(1):212. doi: 10.1186/s12915-025-02314-8. BMC Biol. 2025. PMID: 40660190 Free PMC article.
-
msBERT-Promoter: a multi-scale ensemble predictor based on BERT pre-trained model for the two-stage prediction of DNA promoters and their strengths.BMC Biol. 2024 May 30;22(1):126. doi: 10.1186/s12915-024-01923-z. BMC Biol. 2024. PMID: 38816885 Free PMC article.
-
scRSSL: Residual semi-supervised learning with deep generative models to automatically identify cell types.IET Syst Biol. 2025 Jan-Dec;19(1):e12107. doi: 10.1049/syb2.12107. Epub 2025 Apr 22. IET Syst Biol. 2025. PMID: 40261690 Free PMC article.
References
-
- a) Denisenko E., Guo B. B., Jones M., Hou R., De Kock L., Lassmann T., Poppe D., Clément O., Simmons R. K., Lister R., Forrest A. R. R., Genome biol. 2020, 21, 130; - PMC - PubMed
- b) Kuksin M., Morel D., Aglave M., Danlos F.‐X., Marabelle A., Zinovyev A., Gautheret D., Verlingue L., Eur. J. Cancer 2021, 149, 193. - PubMed
-
- a) Vallania F., Tam A., Lofgren S., Schaffert S., Azad T. D., Bongen E., Haynes W., Alsup M., Alonso M., Davis M., Engleman E., Khatri P., Nat. Commun. 2018, 9, 4735; - PMC - PubMed
- b) Avila Cobos F., Vandesompele J., Mestdagh P., De Preter K., Bioinformatics 2018, 34, 1969; - PubMed
- c) Sturm G., Finotello F., Petitprez F., Zhang J. D., Baumbach J., Fridman W. H., List M., Aneichyk T., Bioinformatics 2019, 35, i436. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
