ProNGF promotes brain metastasis through TrkA/EphA2 induced Src activation in triple negative breast cancer cells
- PMID: 38072918
- PMCID: PMC10710730
- DOI: 10.1186/s40164-023-00463-6
ProNGF promotes brain metastasis through TrkA/EphA2 induced Src activation in triple negative breast cancer cells
Abstract
Background: Triple-Negative Breast Cancer is particularly aggressive, and its metastasis to the brain has a significant psychological impact on patients' quality of life, in addition to reducing survival. The development of brain metastases is particularly harmful in triple-negative breast cancer (TNBC). To date, the mechanisms that induce brain metastasis in TNBC are poorly understood.
Methods: Using a human blood-brain barrier (BBB) in vitro model, an in vitro 3D organotypic extracellular matrix, an ex vivo mouse brain slices co-culture and in an in vivo xenograft experiment, key step of brain metastasis were recapitulated to study TNBC behaviors.
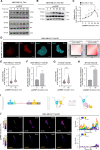
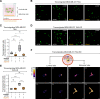
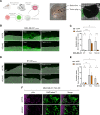
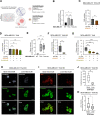
Results: In this study, we demonstrated for the first time the involvement of the precursor of Nerve Growth Factor (proNGF) in the development of brain metastasis. More importantly, our results showed that proNGF acts through TrkA independent of its phosphorylation to induce brain metastasis in TNBC. In addition, we found that proNGF induces BBB transmigration through the TrkA/EphA2 signaling complex. More importantly, our results showed that combinatorial inhibition of TrkA and EphA2 decreased TBNC brain metastasis in a preclinical model.
Conclusions: These disruptive findings provide new insights into the mechanisms underlying brain metastasis with proNGF as a driver of brain metastasis of TNBC and identify TrkA/EphA2 complex as a potential therapeutic target.
Keywords: Brain metastasis; Breast cancer; EphA2; Src; TrkA; proNGF.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Deluche E, Antoine A, Bachelot T, Lardy-Cleaud A, Dieras V, Brain E, Debled M, Jacot W, Mouret-Reynier MA, Goncalves A, Dalenc F, Patsouris A, Ferrero JM, Levy C, Lorgis V, Vanlemmens L, Lefeuvre-Plesse C, Mathoulin-Pelissier S, Petit T, Uwer L, Jouannaud C, Leheurteur M, Lacroix-Triki M, Courtinard C, Perol D, Robain M, Delaloge S. Contemporary outcomes of metastatic breast cancer among 22,000 women from the multicentre ESME cohort 2008–2016. Eur J Cancer. 2020 doi: 10.1016/j.ejca.2020.01.016. - DOI - PubMed
-
- Van’t Veer LJ, Dai H, Van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, Van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002 doi: 10.1038/415530a. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

