Advances in neuroproteomics for neurotrauma: unraveling insights for personalized medicine and future prospects
- PMID: 38073638
- PMCID: PMC10703396
- DOI: 10.3389/fneur.2023.1288740
Advances in neuroproteomics for neurotrauma: unraveling insights for personalized medicine and future prospects
Abstract
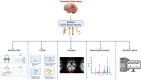
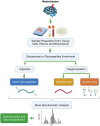
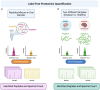
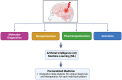
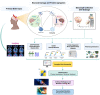
Neuroproteomics, an emerging field at the intersection of neuroscience and proteomics, has garnered significant attention in the context of neurotrauma research. Neuroproteomics involves the quantitative and qualitative analysis of nervous system components, essential for understanding the dynamic events involved in the vast areas of neuroscience, including, but not limited to, neuropsychiatric disorders, neurodegenerative disorders, mental illness, traumatic brain injury, chronic traumatic encephalopathy, and other neurodegenerative diseases. With advancements in mass spectrometry coupled with bioinformatics and systems biology, neuroproteomics has led to the development of innovative techniques such as microproteomics, single-cell proteomics, and imaging mass spectrometry, which have significantly impacted neuronal biomarker research. By analyzing the complex protein interactions and alterations that occur in the injured brain, neuroproteomics provides valuable insights into the pathophysiological mechanisms underlying neurotrauma. This review explores how such insights can be harnessed to advance personalized medicine (PM) approaches, tailoring treatments based on individual patient profiles. Additionally, we highlight the potential future prospects of neuroproteomics, such as identifying novel biomarkers and developing targeted therapies by employing artificial intelligence (AI) and machine learning (ML). By shedding light on neurotrauma's current state and future directions, this review aims to stimulate further research and collaboration in this promising and transformative field.
Keywords: artificial intelligence (AI); machine learning (ML); neuroproteomics; neurotrauma; personalized medicine (PM); proteomics; traumatic brain injuries (TBI).
Copyright © 2023 Kobeissy, Goli, Yadikar, Shakkour, Kurup, Haidar, Alroumi, Mondello, Wang and Mechref.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

