A comprehensive transcriptomic analysis of the bisphenol A affected kidney in mice
- PMID: 38074096
- PMCID: PMC10704486
- DOI: 10.3389/fmolb.2023.1260716
A comprehensive transcriptomic analysis of the bisphenol A affected kidney in mice
Abstract
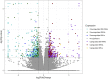
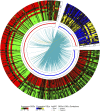
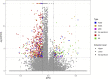
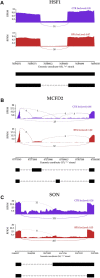
Introduction: Bisphenol A (BPA) is a substance belonging to the endocrine-disrupting chemicals, globally used in the production of polycarbonate plastics. It has been found that BPA enhances carcinogenesis, triggers obesity and exerts a pathogenic effect in several disorders, such as type 2 diabetes, asthma, or increased blood pressure. Recent studies have revealed, that BPA has a harmful impact on the kidneys function, therefore, the current research aimed to explore the specific molecular changes triggered in these organs after oral BPA exposure in mice. Materials and Methods: The experiment was carried out on 12 (3-month-old) female mice. Six mice served as controls. The other 6 mice were treated with BPA in the drinking water at a dose of 50 mg/kg b. w. for 3 months. Then animals were euthanized, the kidneys were collected, and extracted RNA was used to perform RNA-seq. Results: Applied multistep bioinformatics revealed 433 differentially expressed genes (DEGs) in the BPA-treated kidneys (232 upregulated and 201 downregulated). Additionally, 95 differentially expressed long-noncoding RNAs (DELs) were revealed in BPA samples. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations indicated that BPA exposure resulted in profound changes in several essential processes, such as oxidative phosphorylation, mitochondrial and ribosome function, or chemical carcinogenesis. Conclusion: The obtained novel results suggest that BPA has a harmful impact on the fundamental processes of the kidney and significantly impairs its function by inducing mitochondrial dysfunction leading to oxidative stress and reactive oxygen species production.
Keywords: RNA-Seq; bisphenol A; kidney; mitochondrial dysfunction; oxidative stress.
Copyright © 2023 Wiszpolska, Lepiarczyk, Paukszto, Makowczenko, Lipka, Maździarz, Polak, Makowska, Gonkowski, Correia-de-Sá and Majewska.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
-
- AACR Project GENIE Consortium (2017). AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 7, 818–831. 10.1158/2159-8290.CD-17-0151 - DOI - PMC - PubMed
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
LinkOut - more resources
Full Text Sources

