A toolbox of machine learning software to support microbiome analysis
- PMID: 38075858
- PMCID: PMC10704913
- DOI: 10.3389/fmicb.2023.1250806
A toolbox of machine learning software to support microbiome analysis
Abstract
The human microbiome has become an area of intense research due to its potential impact on human health. However, the analysis and interpretation of this data have proven to be challenging due to its complexity and high dimensionality. Machine learning (ML) algorithms can process vast amounts of data to uncover informative patterns and relationships within the data, even with limited prior knowledge. Therefore, there has been a rapid growth in the development of software specifically designed for the analysis and interpretation of microbiome data using ML techniques. These software incorporate a wide range of ML algorithms for clustering, classification, regression, or feature selection, to identify microbial patterns and relationships within the data and generate predictive models. This rapid development with a constant need for new developments and integration of new features require efforts into compile, catalog and classify these tools to create infrastructures and services with easy, transparent, and trustable standards. Here we review the state-of-the-art for ML tools applied in human microbiome studies, performed as part of the COST Action ML4Microbiome activities. This scoping review focuses on ML based software and framework resources currently available for the analysis of microbiome data in humans. The aim is to support microbiologists and biomedical scientists to go deeper into specialized resources that integrate ML techniques and facilitate future benchmarking to create standards for the analysis of microbiome data. The software resources are organized based on the type of analysis they were developed for and the ML techniques they implement. A description of each software with examples of usage is provided including comments about pitfalls and lacks in the usage of software based on ML methods in relation to microbiome data that need to be considered by developers and users. This review represents an extensive compilation to date, offering valuable insights and guidance for researchers interested in leveraging ML approaches for microbiome analysis.
Keywords: data integration; feature analysis; feature generation; machine learning; microbial gene prediction; microbial metabolic modeling; microbiome; software.
Copyright © 2023 Marcos-Zambrano, López-Molina, Bakir-Gungor, Frohme, Karaduzovic-Hadziabdic, Klammsteiner, Ibrahimi, Lahti, Loncar Turukalo, Dhamo, Simeon, Nechyporenko, Pio, Przymus, Sampri, Trajkovik, Lacruz-Pleguezuelos, Aasmets, Araujo, Anagnostopoulos, Aydemir, Berland, Calle, Ceci, Duman, Gündoğdu, Havulinna, Kaka Bra, Kalluci, Karav, Lode, Lopes, May, Nap, Nedyalkova, Paciência, Pasic, Pujolassos, Shigdel, Susín, Thiele, Truică, Wilmes, Yilmaz, Yousef, Claesson, Truu, Carrillo de Santa Pau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
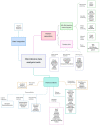
Figures

References
-
- Adapsyn Bioscience (2022). Available at: https://adapsyn.com/.
Publication types
LinkOut - more resources
Full Text Sources

