Thiocillin contributes to the ecological fitness of Bacillus cereus ATCC 14579 during interspecies interactions with Myxococcus xanthus
- PMID: 38075900
- PMCID: PMC10704990
- DOI: 10.3389/fmicb.2023.1295262
Thiocillin contributes to the ecological fitness of Bacillus cereus ATCC 14579 during interspecies interactions with Myxococcus xanthus
Abstract
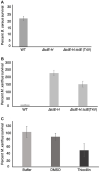
The soil-dwelling delta-proteobacterium Myxococcus xanthus is a model organism to study predation and competition. M. xanthus preys on a broad range of bacteria mediated by lytic enzymes, exopolysaccharides, Type-IV pilus-based motility, and specialized metabolites. Competition between M. xanthus and prey bacterial strains with various specialized metabolite profiles indicates a range of fitness, suggesting that specialized metabolites contribute to prey survival. To expand our understanding of how specialized metabolites affect predator-prey dynamics, we assessed interspecies interactions between M. xanthus and two strains of Bacillus cereus. While strain ATCC 14579 resisted predation, strain T was found to be highly sensitive to M. xanthus predation. The interaction between B. cereus ATCC 14579 and M. xanthus appears to be competitive, resulting in population loss for both predator and prey. Genome analysis revealed that ATCC 14579 belongs to a clade that possesses the biosynthetic gene cluster for production of thiocillins, whereas B. cereus strain T lacks those genes. Further, purified thiocillin protects B. cereus strains unable to produce this specialized metabolite, strengthening the finding that thiocillin protects against predation and contributes to the ecological fitness of B. cereus ATCC 14579. Lastly, strains that produce thiocillin appear to confer some level of protection to their own antibiotic by encoding an additional copy of the L11 ribosomal protein, a known target for thiopeptides. This work highlights the importance of specialized metabolites affecting predator-prey dynamics in soil microenvironments.
Keywords: B. cereus; M. xanthus; competition; predator–prey interactions; specialized metabolites.
Copyright © 2023 Müller, DeLeon, Atkinson, Saravia, Kellogg, Shank and Kirby.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identification of Functions Affecting Predator-Prey Interactions between Myxococcus xanthus and Bacillus subtilis.J Bacteriol. 2016 Nov 18;198(24):3335-3344. doi: 10.1128/JB.00575-16. Print 2016 Dec 15. J Bacteriol. 2016. PMID: 27698086 Free PMC article.
-
Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus.Appl Environ Microbiol. 2014 Sep;80(18):5603-10. doi: 10.1128/AEM.01621-14. Epub 2014 Jul 7. Appl Environ Microbiol. 2014. PMID: 25002419 Free PMC article.
-
Dynamics of Solitary Predation by Myxococcus xanthus on Escherichia coli Observed at the Single-Cell Level.Appl Environ Microbiol. 2020 Jan 21;86(3):e02286-19. doi: 10.1128/AEM.02286-19. Print 2020 Jan 21. Appl Environ Microbiol. 2020. PMID: 31704687 Free PMC article.
-
The Genetics of Prey Susceptibility to Myxobacterial Predation: A Review, Including an Investigation into Pseudomonas aeruginosa Mutations Affecting Predation by Myxococcus xanthus.Microb Physiol. 2021;31(2):57-66. doi: 10.1159/000515546. Epub 2021 Apr 1. Microb Physiol. 2021. PMID: 33794538 Review.
-
Myxococcus xanthus predation: an updated overview.Front Microbiol. 2024 Jan 24;15:1339696. doi: 10.3389/fmicb.2024.1339696. eCollection 2024. Front Microbiol. 2024. PMID: 38328431 Free PMC article. Review.
Cited by
-
Reutericyclin, a specialized metabolite of Limosilactobacillus reuteri, mitigates risperidone-induced weight gain in mice.Gut Microbes. 2025 Dec;17(1):2477819. doi: 10.1080/19490976.2025.2477819. Epub 2025 Apr 7. Gut Microbes. 2025. PMID: 40190120 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

