Analysis of gene expression profile for identification of novel gene signatures during dengue infection
- PMID: 38076406
- PMCID: PMC10699721
- DOI: 10.1016/j.imj.2023.02.002
Analysis of gene expression profile for identification of novel gene signatures during dengue infection
Abstract
Background: Dengue is a major arthropod-borne viral disease spreading rapidly across the globe. The absence of vaccines and inadequate vector control measures leads to further expansion of dengue in many regions globally. Hence, the identification of genes involved in the pathogenesis of dengue will help to understand the molecular basis of the disease and the genes responsible for the disease progression.
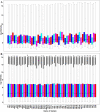
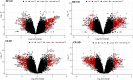
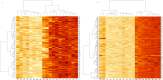
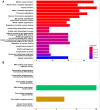
Methods: In the present study, a meta-analysis was carried out using dengue gene expression data obtained from Gene Expression Omnibus repository. The differentially expressed genes such as CCNB1 and CCNB2 (G2/mitotic-specific cyclin-B2 and B1) were upregulated in dengue fever to control (DF-CO) and severe dengue (dengue hemorrhagic fever [DHF]) to control (DHF-CO) were identified as key genes for controlling the major pathways (cell cycle, oocyte meiosis, p53 signaling pathway, cellular senescence and progesterone-mediated oocyte maturation). Similarly, interferon alpha-inducible (IFI27) genes, type-I and type-III interferon (IFN) signaling genes (STAT1 and STAT2), B cell activation and survival genes (TNFSF13B, TNFRSF17) and toll like receptor (TLR7) genes were differentially up activated during DF-CO and DHF-CO. Followed by, Cytoscape was used to identify the immune system process and topological analysis.
Results: The results showed that the top differentially expressed genes under the statistical significance p <0.001, which is majorly involved in Kyoto Encyclopedia of Genes and Genomes orthology K05868 and K21770 with gene names CCNB1 and CCNB2. In addition to this, the immune system profile showed up-regulation of IL12A, CXCR3, TNFSF13B, IFI27, TNFRSF17, STAT, STAT2, and TLR7 genes in DF-CO and DHF-CO act as immunological signatures for inducing the immune response towards dengue infection.
Conclusions: The current study could aid in understanding of molecular pathogenesis, genes and corresponding pathway upon dengue infection, and could facilitate for identification of novel drug targets and prognostic markers.
Keywords: Data analysis; Dengue fever; Gene expression; Gene signatures; Microarray; Severe dengue.
© 2023 The Author(s).
Figures


References
-
- Arunachalam N., Murty U.S., Kabilan L., et al. Studies on dengue in rural areas of Kurnool District, Andhra Pradesh, India. J. Am. Mosq. Control Assoc. 2004;20(1):87–90. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

