This is a preprint.
Translation elongation as a rate limiting step of protein production
- PMID: 38076849
- PMCID: PMC10705293
- DOI: 10.1101/2023.11.27.568910
Translation elongation as a rate limiting step of protein production
Abstract
The impact of synonymous codon choice on protein output has important implications for understanding endogenous gene expression and design of synthetic mRNAs. Synonymous codons are decoded at different speeds, but simple models predict that this should not drive protein output. Instead, translation initiation should be the rate limiting step for production of protein per mRNA, with little impact of codon choice. Previously, we used a neural network model to design a series of synonymous fluorescent reporters and showed that their protein output in yeast spanned a seven-fold range corresponding to their predicted translation elongation speed. Here, we show that this effect is not due primarily to the established impact of slow elongation on mRNA stability, but rather, that slow elongation further decreases the number of proteins made per mRNA. We combine simulations and careful experiments on fluorescent reporters to show that translation is limited on non-optimally encoded transcripts. Using a genome-wide CRISPRi screen, we find that impairing translation initiation attenuates the impact of slow elongation, showing a dynamic balance between rate limiting steps of protein production. Our results show that codon choice can directly limit protein production across the full range of endogenous variability in codon usage.
Conflict of interest statement
Declaration of interests N.T.I. is a shareholder of Velia Therapeutics and a shareholder and member of the scientific advisory board of Tevard Biosciences. L.F.L. holds a patent on the iXnos mRNA design method used here.
Figures

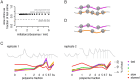
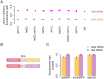


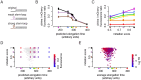
References
-
- Bazzini Ariel A., Del Viso Florencia, Moreno-Mateos Miguel A., Johnstone Timothy G., Vejnar Charles E., Qin Yidan, Yao Jun, Khokha Mustafa K., and Giraldez Antonio J.. 2016. “Codon Identity Regulates mRNA Stability and Translation Efficiency during the Maternal-to-Zygotic Transition.” The EMBO Journal 35 (19): 2087–2103. - PMC - PubMed
References (Methods)
-
- Ares Manuel. 2012. “Isolation of Total RNA from Yeast Cell Cultures.” Cold Spring Harbor Protocols 2012 (10): 1082–86. - PubMed
-
- Gietz R. Daniel, and Woods Robin A.. 2002. “Transformation of Yeast by Lithium Acetate/single-Stranded Carrier DNA/polyethylene Glycol Method.” Methods in Enzymology 350: 87–96. - PubMed
-
- Gillespie Daniel T. 1976. “A General Method for Numerically Simulating the Stochastic Time Evolution of Coupled Chemical Reactions.” Journal of Computational Physics 22 (4): 403–34.
-
- McGeachy Anna M., Meacham Zuriah A., and Ingolia Nicholas T.. 2019. “An Accessible Continuous-Culture Turbidostat for Pooled Analysis of Complex Libraries.” ACS Synthetic Biology 8 (4): 844–56. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
