Processing of matched and mismatched rNMPs in DNA by archaeal ribonucleotide excision repair
- PMID: 38077150
- PMCID: PMC10698281
- DOI: 10.1016/j.isci.2023.108479
Processing of matched and mismatched rNMPs in DNA by archaeal ribonucleotide excision repair
Abstract
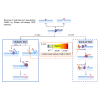
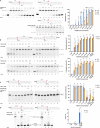
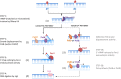
Ribonucleoside monophosphates (rNMPs) are the main non-canonical nucleotides in genomic DNA, and their incorporation can occur as mismatches or matches in vivo. To counteract the mutagenic potential of rNMPs in DNA, all organisms evolved ribonucleotide excision repair (RER), a mechanism initiated by type 2 RNase H. Here, we describe the in vitro reconstitution of matched and mismatched rNMP repair using archaeal RER enzymes. Our data suggest two types of RER pathways, including the classical flap RER and a backup RER with the order of reactions changed for Fen1 and Pols. The genomic rNMP level in RER-deficient or PolB-deficient archaeal cells along with in vitro reconstitution of RER suggests an in vivo role of PolD in RER. Our results provide insights into how matched and mismatched rNMPs may be processed by RER.
Keywords: Bacteriology; Biochemistry; Molecular biology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
High density of unrepaired genomic ribonucleotides leads to Topoisomerase 1-mediated severe growth defects in absence of ribonucleotide reductase.Nucleic Acids Res. 2020 May 7;48(8):4274-4297. doi: 10.1093/nar/gkaa103. Nucleic Acids Res. 2020. PMID: 32187369 Free PMC article.
-
Unlike the Escherichia coli counterpart, archaeal RNase HII cannot process ribose monophosphate abasic sites and oxidized ribonucleotides embedded in DNA.J Biol Chem. 2019 Aug 30;294(35):13061-13072. doi: 10.1074/jbc.RA119.009493. Epub 2019 Jul 12. J Biol Chem. 2019. PMID: 31300556 Free PMC article.
-
Mutagenic cost of ribonucleotides in bacterial DNA.Proc Natl Acad Sci U S A. 2017 Oct 31;114(44):11733-11738. doi: 10.1073/pnas.1710995114. Epub 2017 Oct 16. Proc Natl Acad Sci U S A. 2017. PMID: 29078353 Free PMC article.
-
Current perspectives on mechanisms of ribonucleotide incorporation and processing in mammalian DNA.Genes Environ. 2019 Jan 25;41:3. doi: 10.1186/s41021-019-0118-7. eCollection 2019. Genes Environ. 2019. PMID: 30700998 Free PMC article. Review.
-
Molecular and physiological consequences of faulty eukaryotic ribonucleotide excision repair.EMBO J. 2020 Feb 3;39(3):e102309. doi: 10.15252/embj.2019102309. Epub 2019 Dec 12. EMBO J. 2020. PMID: 31833079 Free PMC article. Review.
References
-
- Lemor M., Kong Z., Henry E., Brizard R., Laurent S., Bossé A., Henneke G. Differential Activities of DNA Polymerases in Processing Ribonucleotides during DNA Synthesis in Archaea. J. Mol. Biol. 2018;430:4908–4924. - PubMed
-
- Rumbaugh J.A., Murante R.S., Shi S., Bambara R.A. Creation and removal of embedded ribonucleotides in chromosomal DNA during mammalian Okazaki fragment processing. J. Biol. Chem. 1997;272:22591–22599. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

