High-dimensional phenotyping of the peripheral immune response in community-acquired pneumonia
- PMID: 38077404
- PMCID: PMC10704504
- DOI: 10.3389/fimmu.2023.1260283
High-dimensional phenotyping of the peripheral immune response in community-acquired pneumonia
Abstract
Background: Community-acquired pneumonia (CAP) represents a major health burden worldwide. Dysregulation of the immune response plays an important role in adverse outcomes in patients with CAP.
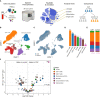
Methods: We analyzed peripheral blood mononuclear cells by 36-color spectral flow cytometry in adult patients hospitalized for CAP (n=40), matched control subjects (n=31), and patients hospitalized for COVID-19 (n=35).
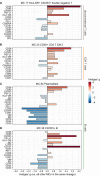
Results: We identified 86 immune cell metaclusters, 19 of which (22.1%) were differentially abundant in patients with CAP versus matched controls. The most notable differences involved classical monocyte metaclusters, which were more abundant in CAP and displayed phenotypic alterations reminiscent of immunosuppression, increased susceptibility to apoptosis, and enhanced expression of chemokine receptors. Expression profiles on classical monocytes, driven by CCR7 and CXCR5, divided patients with CAP into two clusters with a distinct inflammatory response and disease course. The peripheral immune response in patients with CAP was highly similar to that in patients with COVID-19, but increased CCR7 expression on classical monocytes was only present in CAP.
Conclusion: CAP is associated with profound cellular changes in blood that mainly relate to classical monocytes and largely overlap with the immune response detected in COVID-19.
Keywords: COVID-19; chemokine receptors; host response; immunophenotyping; immunosuppression; monocytes; pneumonia; spectral flow cytometry.
Copyright © 2023 Reijnders, Schuurman, Verhoeff, van den Braber, Douma, Faber, Paul, Wiersinga, Saris, Garcia Vallejo and van der Poll.
Conflict of interest statement
Author AP was employed by the company Cytek Biosciences, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Abbafati C, Abbas KM, Abbasi-Kangevari M, Abd-Allah F, Abdelalim A, Abdollahi M, et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet (2020) 396:1204–22. doi: 10.1016/S0140-6736(20)30925-9 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

