Fibril structures of TFG protein mutants validate the identification of TFG as a disease-related amyloid protein by the IMPAcT method
- PMID: 38077690
- PMCID: PMC10703350
- DOI: 10.1093/pnasnexus/pgad402
Fibril structures of TFG protein mutants validate the identification of TFG as a disease-related amyloid protein by the IMPAcT method
Abstract
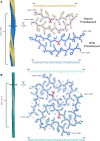
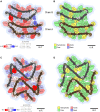
We previously presented a bioinformatic method for identifying diseases that arise from a mutation in a protein's low-complexity domain that drives the protein into pathogenic amyloid fibrils. One protein so identified was the tropomyosin-receptor kinase-fused gene protein (TRK-fused gene protein or TFG). Mutations in TFG are associated with degenerative neurological conditions. Here, we present experimental evidence that confirms our prediction that these conditions are amyloid-related. We find that the low-complexity domain of TFG containing the disease-related mutations G269V or P285L forms amyloid fibrils, and we determine their structures using cryo-electron microscopy (cryo-EM). These structures are unmistakably amyloid in nature and confirm the propensity of the mutant TFG low-complexity domain to form amyloid fibrils. Also, despite resulting from a pathogenic mutation, the fibril structures bear some similarities to other amyloid structures that are thought to be nonpathogenic and even functional, but there are other factors that support these structures' relevance to disease, including an increased propensity to form amyloid compared with the wild-type sequence, structure-stabilizing influence from the mutant residues themselves, and double-protofilament amyloid cores. Our findings elucidate two potentially disease-relevant structures of a previously unknown amyloid and also show how the structural features of pathogenic amyloid fibrils may not conform to the features commonly associated with pathogenicity.
Keywords: Charcot–Marie–Tooth disease; amyloid; cryo-electron microscopy; hereditary motor and sensory neuropathy with proximal dominant involvement; mutation.
© The Author(s) 2023. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures



Similar articles
-
Bioinformatic identification of previously unrecognized amyloidogenic proteins.J Biol Chem. 2022 May;298(5):101920. doi: 10.1016/j.jbc.2022.101920. Epub 2022 Apr 9. J Biol Chem. 2022. PMID: 35405097 Free PMC article.
-
TFG mutation induces haploinsufficiency and drives axonal Charcot-Marie-Tooth disease by causing neurite degeneration.CNS Neurosci Ther. 2022 Dec;28(12):2076-2089. doi: 10.1111/cns.13943. Epub 2022 Aug 19. CNS Neurosci Ther. 2022. PMID: 35986567 Free PMC article.
-
Continuum of phenotypes in hereditary motor and sensory neuropathy with proximal predominance and Charcot-Marie-Tooth patients with TFG mutation.Am J Med Genet A. 2019 Aug;179(8):1507-1515. doi: 10.1002/ajmg.a.61184. Epub 2019 May 20. Am J Med Genet A. 2019. PMID: 31111683
-
TFG-Related Neurologic Disorders: New Insights Into Relationships Between Endoplasmic Reticulum and Neurodegeneration.J Neuropathol Exp Neurol. 2016 Apr;75(4):299-305. doi: 10.1093/jnen/nlw009. Epub 2016 Mar 4. J Neuropathol Exp Neurol. 2016. PMID: 26945032 Review.
-
Targeting tropomyosin-receptor kinase fused gene in cancer.Anticancer Res. 2014 Apr;34(4):1595-600. Anticancer Res. 2014. PMID: 24692687 Review.
References
-
- Wootton JC, Federhen S. 1993. Statistics of local complexity in amino acid sequences and sequence databases. Comput Chem. 17:149–163.
-
- Tsai P-C, et al. 2014. A novel TFG mutation causes Charcot-Marie-Tooth disease type 2 and impairs TFG function. Neurology. 83:903–912. - PubMed

