Main and papain-like proteases as prospective targets for pharmacological treatment of coronavirus SARS-CoV-2
- PMID: 38077980
- PMCID: PMC10698513
- DOI: 10.1039/d3ra06479d
Main and papain-like proteases as prospective targets for pharmacological treatment of coronavirus SARS-CoV-2
Abstract
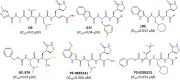
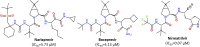
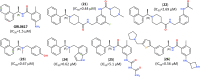
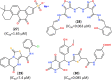
The pandemic caused by the coronavirus SARS-CoV-2 led to a global crisis in the world healthcare system. Despite some progress in the creation of antiviral vaccines and mass vaccination of the population, the number of patients continues to grow because of the spread of new SARS-CoV-2 mutations. There is an urgent need for direct-acting drugs capable of suppressing or stopping the main mechanisms of reproduction of the coronavirus SARS-CoV-2. Several studies have shown that the successful replication of the virus in the cell requires proteolytic cleavage of the protein structures of the virus. Two proteases are crucial in replicating SARS-CoV-2 and other coronaviruses: the main protease (Mpro) and the papain-like protease (PLpro). In this review, we summarize the essential viral proteins of SARS-CoV-2 required for its viral life cycle as targets for chemotherapy of coronavirus infection and provide a critical summary of the development of drugs against COVID-19 from the drug repurposing strategy up to the molecular design of novel covalent and non-covalent agents capable of inhibiting virus replication. We overview the main antiviral strategy and the choice of SARS-CoV-2 Mpro and PLpro proteases as promising targets for pharmacological impact on the coronavirus life cycle.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


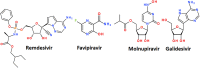

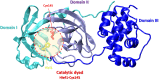
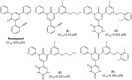
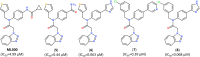









References
-
- Gil C. Ginex T. Maestro I. Nozal V. Barrado-Gil L. Cuesta-Geijo M. Á. Urquiza J. Ramírez D. Alonso C. Campillo N. E. Martinez A. COVID-19: Drug Targets and Potential Treatments. J. Med. Chem. 2020;63:12359–12386. - PubMed
-
- Al-Shargi O. Y. Alzaid S. M. Al-Dosari B. S. Alzubaidi A. S. Adverse events associated with COVID-19 treatment and their possible relationship with patient characteristics: A narrative review. J. Appl. Pharm. Sci. 2023;13:41–49.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

