Nonuniversal impact of cholesterol on membranes mobility, curvature sensing and elasticity
- PMID: 38081812
- PMCID: PMC10713574
- DOI: 10.1038/s41467-023-43892-x
Nonuniversal impact of cholesterol on membranes mobility, curvature sensing and elasticity
Abstract
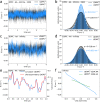
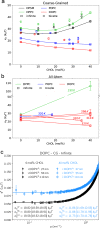
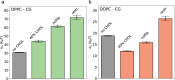
Biological membranes, composed mainly of phospholipids and cholesterol, play a vital role as cellular barriers. They undergo localized reshaping in response to environmental cues and protein interactions, with the energetics of deformations crucial for exerting biological functions. This study investigates the non-universal role of cholesterol on the structure and elasticity of saturated and unsaturated lipid membranes. Our study uncovers a highly cooperative relationship between thermal membrane bending and local cholesterol redistribution, with cholesterol showing a strong preference for the compressed membrane leaflet. Remarkably, in unsaturated membranes, increased cholesterol mobility enhances cooperativity, resulting in membrane softening despite membrane thickening and lipid compression caused by cholesterol. These findings elucidate the intricate interplay between thermodynamic forces and local molecular interactions that govern collective properties of membranes.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

