Conformational plasticity of RAS Q61 family of neoepitopes results in distinct features for targeted recognition
- PMID: 38081856
- PMCID: PMC10713829
- DOI: 10.1038/s41467-023-43654-9
Conformational plasticity of RAS Q61 family of neoepitopes results in distinct features for targeted recognition
Abstract
The conformational landscapes of peptide/human leucocyte antigen (pHLA) protein complexes encompassing tumor neoantigens provide a rationale for target selection towards autologous T cell, vaccine, and antibody-based therapeutic modalities. Here, using complementary biophysical and computational methods, we characterize recurrent RAS55-64 Q61 neoepitopes presented by the common HLA-A*01:01 allotype. We integrate sparse NMR restraints with Rosetta docking to determine the solution structure of NRASQ61K/HLA-A*01:01, which enables modeling of other common RAS55-64 neoepitopes. Hydrogen/deuterium exchange mass spectrometry experiments alongside molecular dynamics simulations reveal differences in solvent accessibility and conformational plasticity across a panel of common Q61 neoepitopes that are relevant for recognition by immunoreceptors. Finally, we predict binding and provide structural models of NRASQ61K antigens spanning the entire HLA allelic landscape, together with in vitro validation for HLA-A*01:191, HLA-B*15:01, and HLA-C*08:02. Our work provides a basis to delineate the solution surface features and immunogenicity of clinically relevant neoepitope/HLA targets for cancer therapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
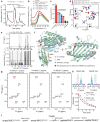



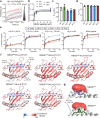


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

