CSGALNACT2 restricts ovarian cancer migration and invasion by modulating MAPK/ERK pathway through DUSP1
- PMID: 38082211
- PMCID: PMC11219422
- DOI: 10.1007/s13402-023-00903-9
CSGALNACT2 restricts ovarian cancer migration and invasion by modulating MAPK/ERK pathway through DUSP1
Abstract
Purpose: Ovarian cancer is one of the leading causes of cancer-related death among women. CSGALNACT2 is a vital Golgi transferase and is related to a variety of human diseases. However, its expression pattern and function in ovarian cancer remain uncertain.
Methods: The Cancer Genome Atlas and GEPIA databases were used to assess the expression of CSGALNACT2 in ovarian cancer patients. RNA-seq, qRT-PCR, and IHC were used to verify the expression of CSGALNACT2 in ovarian cancer tissues. Then, in vivo and in vitro experiments were conducted to evaluate the role of CSGALNACT2 in the progression of ovarian cancer. RNA-seq and GSEA were used to reveal the potential biological function and oncogenic pathways of CSGALNACT2.
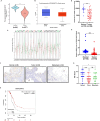
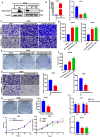
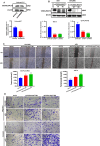
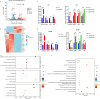
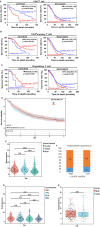
Results: We demonstrated that the mRNA expression and protein level of CSGALNACT2 were significantly downregulated in ovarian cancer and ovarian cancer metastatic tissues. CSGALNACT2 can significantly inhibit the migration, invasion, and clonogenic growth of ovarian cancer in vitro and is progressively lost during ovarian cancer progression in vivo. CSGALNACT2 suppresses ovarian cancer migration and invasion via DUSP1 modulation of the MAPK/ERK pathway through RNA-seq, KEGG analysis, and Western blotting. Moreover, CSGALNACT2 expression was correlated with immune cell infiltration and had prognostic value in different immune cell-enriched or decreased ovarian cancer. In addition, patients with CSGALNACT2 downregulation are less likely to benefit from immunotherapy.
Conclusion: As an ovarian cancer suppressor gene, CSGALNACT2 inhibits the development of ovarian cancer, and it might be used as a prognostic biomarker in patients with ovarian cancer.
Keywords: CSGALNACT2; DUSP1; MAPK/ERK pathway; Migration and invasion; Ovarian cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures





References
-
- Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11(4):933–959. doi: 10.1158/2159-8290.CD-20-1808. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

