Acute ischemia induces spatially and transcriptionally distinct microglial subclusters
- PMID: 38082331
- PMCID: PMC10712107
- DOI: 10.1186/s13073-023-01257-5
Acute ischemia induces spatially and transcriptionally distinct microglial subclusters
Abstract
Background: Damage in the ischemic core and penumbra after stroke affects patient prognosis. Microglia immediately respond to ischemic insult and initiate immune inflammation, playing an important role in the cellular injury after stroke. However, the microglial heterogeneity and the mechanisms involved remain unclear.
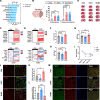
Methods: We first performed single-cell RNA-sequencing (scRNA-seq) and spatial transcriptomics (ST) on middle cerebral artery occlusion (MCAO) mice from three time points to determine stroke-associated microglial subclusters and their spatial distributions. Furthermore, the expression of microglial subcluster-specific marker genes and the localization of different microglial subclusters were verified on MCAO mice through RNAscope and immunofluorescence. Gene set variation analysis (GSVA) was performed to reveal functional characteristics of microglia sub-clusters. Additionally, ingenuity pathway analysis (IPA) was used to explore upstream regulators of microglial subclusters, which was confirmed by immunofluorescence, RT-qPCR, shRNA-mediated knockdown, and targeted metabolomics. Finally, the infarct size, neurological deficits, and neuronal apoptosis were evaluated in MCAO mice after manipulation of specific microglial subcluster.
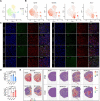
Results: We discovered stroke-associated microglial subclusters in the brains of MCAO mice. We also identified novel marker genes of these microglial subclusters and defined these cells as ischemic core-associated (ICAM) and ischemic penumbra-associated (IPAM) microglia, according to their spatial distribution. ICAM, induced by damage-associated molecular patterns, are probably fueled by glycolysis, and exhibit increased pro-inflammatory cytokines and chemokines production. BACH1 is a key transcription factor driving ICAM generation. In contrast, glucocorticoids, which are enriched in the penumbra, likely trigger IPAM formation, which are presumably powered by the citrate cycle and oxidative phosphorylation and are characterized by moderate pro-inflammatory responses, inflammation-alleviating metabolic features, and myelinotrophic properties.
Conclusions: ICAM could induce excessive neuroinflammation, aggravating brain injury, whereas IPAM probably exhibit neuroprotective features, which could be essential for the homeostasis and survival of cells in the penumbra. Our findings provide a biological basis for targeting specific microglial subclusters as a potential therapeutic strategy for ischemic stroke.
Keywords: BACH1; Glucocorticoids; Ischemic stroke; Microglia; Spatial transcriptomics; scRNA-seq.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

