A prophage encoded ribosomal RNA methyltransferase regulates the virulence of Shiga-toxin-producing Escherichia coli (STEC)
- PMID: 38084890
- PMCID: PMC10810198
- DOI: 10.1093/nar/gkad1150
A prophage encoded ribosomal RNA methyltransferase regulates the virulence of Shiga-toxin-producing Escherichia coli (STEC)
Abstract
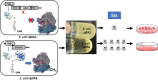
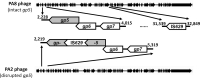
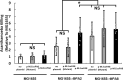
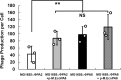
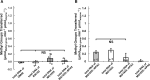
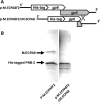
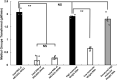
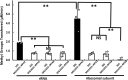
Shiga toxin (Stx) released by Shiga toxin producing Escherichia coli (STEC) causes life-threatening illness. Its production and release require induction of Stx-encoding prophage resident within the STEC genome. We identified two different STEC strains, PA2 and PA8, bearing Stx-encoding prophage whose sequences primarily differ by the position of an IS629 insertion element, yet differ in their abilities to kill eukaryotic cells and whose prophages differ in their spontaneous induction frequencies. The IS629 element in ϕPA2, disrupts an ORF predicted to encode a DNA adenine methyltransferase, whereas in ϕPA8, this element lies in an intergenic region. Introducing a plasmid expressing the methyltransferase gene product into ϕPA2 bearing-strains increases both the prophage spontaneous induction frequency and virulence to those exhibited by ϕPA8 bearing-strains. However, a plasmid bearing mutations predicted to disrupt the putative active site of the methyltransferase does not complement either of these defects. When complexed with a second protein, the methyltransferase holoenzyme preferentially uses 16S rRNA as a substrate. The second subunit is responsible for directing the preferential methylation of rRNA. Together these findings reveal a previously unrecognized role for rRNA methylation in regulating induction of Stx-encoding prophage.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Differential induction of Shiga toxin in environmental Escherichia coli O145:H28 strains carrying the same genotype as the outbreak strains.Int J Food Microbiol. 2021 Feb 2;339:109029. doi: 10.1016/j.ijfoodmicro.2020.109029. Epub 2020 Dec 23. Int J Food Microbiol. 2021. PMID: 33360585
-
DNA adenine methylase, not the PstI restriction-modification system, regulates virulence gene expression in Shiga toxin-producing Escherichia coli.Food Microbiol. 2021 Jun;96:103722. doi: 10.1016/j.fm.2020.103722. Epub 2020 Dec 31. Food Microbiol. 2021. PMID: 33494894
-
Induction of Shiga Toxin-Encoding Prophage by Abiotic Environmental Stress in Food.Appl Environ Microbiol. 2017 Sep 15;83(19):e01378-17. doi: 10.1128/AEM.01378-17. Print 2017 Oct 1. Appl Environ Microbiol. 2017. PMID: 28778890 Free PMC article.
-
Shiga toxins and stx phages: highly diverse entities.Microbiology (Reading). 2015 Mar;161(Pt 3):451-62. doi: 10.1099/mic.0.000003. Epub 2014 Dec 5. Microbiology (Reading). 2015. PMID: 25479836 Review.
-
Altruism of Shiga toxin-producing Escherichia coli: recent hypothesis versus experimental results.Front Cell Infect Microbiol. 2013 Jan 4;2:166. doi: 10.3389/fcimb.2012.00166. eCollection 2012. Front Cell Infect Microbiol. 2013. PMID: 23316482 Free PMC article. Review.
Cited by
-
Gut phageome: challenges in research and impact on human microbiota.Front Microbiol. 2024 Mar 22;15:1379382. doi: 10.3389/fmicb.2024.1379382. eCollection 2024. Front Microbiol. 2024. PMID: 38585689 Free PMC article. Review.
References
-
- Frenzen P.D., Drake A., Angulo F.J., Group E.I.P.F.W.. Economic cost of illness due to Escherichia coli O157 infections in the United States. J. Food Prot. 2005; 68:2623–2630. - PubMed
-
- Karmali M.A. Emerging public health challenges of Shiga toxin–producing Escherichia coli related to changes in the pathogen, the population, and the environment. Clin. Infect. Dis. 2017; 64:371–376. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

