Commensal bacteria promote type I interferon signaling to maintain immune tolerance in mice
- PMID: 38085267
- PMCID: PMC10716256
- DOI: 10.1084/jem.20230063
Commensal bacteria promote type I interferon signaling to maintain immune tolerance in mice
Abstract
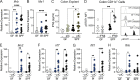
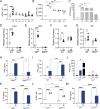
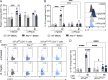
Type I interferons (IFNs) exert a broad range of biological effects important in coordinating immune responses, which have classically been studied in the context of pathogen clearance. Yet, whether immunomodulatory bacteria operate through IFN pathways to support intestinal immune tolerance remains elusive. Here, we reveal that the commensal bacterium, Bacteroides fragilis, utilizes canonical antiviral pathways to modulate intestinal dendritic cells (DCs) and regulatory T cell (Treg) responses. Specifically, IFN signaling is required for commensal-induced tolerance as IFNAR1-deficient DCs display blunted IL-10 and IL-27 production in response to B. fragilis. We further establish that IFN-driven IL-27 in DCs is critical in shaping the ensuing Foxp3+ Treg via IL-27Rα signaling. Consistent with these findings, single-cell RNA sequencing of gut Tregs demonstrated that colonization with B. fragilis promotes a distinct IFN gene signature in Foxp3+ Tregs during intestinal inflammation. Altogether, our findings demonstrate a critical role of commensal-mediated immune tolerance via tonic type I IFN signaling.
© 2023 Vasquez Ayala et al.
Conflict of interest statement
Disclosures: L.-F. Lu reported personal fees from Elixiron Immunotherapeutics and grants from Molecular Axiom outside the submitted work. No other disclosures were reported.
Figures






References
-
- Abt, M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J., et al. . 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 37:158–170. 10.1016/j.immuni.2012.04.011 - DOI - PMC - PubMed

