Cortical somatostatin long-range projection neurons and interneurons exhibit divergent developmental trajectories
- PMID: 38086373
- PMCID: PMC7618239
- DOI: 10.1016/j.neuron.2023.11.013
Cortical somatostatin long-range projection neurons and interneurons exhibit divergent developmental trajectories
Abstract
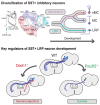
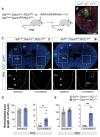
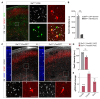
The mammalian cerebral cortex contains an extraordinary diversity of cell types that emerge by implementing different developmental programs. Delineating when and how cellular diversification occurs is particularly challenging for cortical inhibitory neurons because they represent a small proportion of all cortical cells and have a protracted development. Here, we combine single-cell RNA sequencing and spatial transcriptomics to characterize the emergence of neuronal diversity among somatostatin-expressing (SST+) cells in mice. We found that SST+ inhibitory neurons segregate during embryonic stages into long-range projection (LRP) neurons and two types of interneurons, Martinotti cells and non-Martinotti cells, following distinct developmental trajectories. Two main subtypes of LRP neurons and several subtypes of interneurons are readily distinguishable in the embryo, although interneuron diversity is likely refined during early postnatal life. Our results suggest that the timing for cellular diversification is unique for different subtypes of SST+ neurons and particularly divergent for LRP neurons and interneurons.
Keywords: GABA; cell fate; cerebral cortex; development; interneuron; mouse; somatostatin; specification.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

