The AP-2 complex interacts with γ-TuRC and regulates the proliferative capacity of neural progenitors
- PMID: 38086550
- PMCID: PMC10716017
- DOI: 10.26508/lsa.202302029
The AP-2 complex interacts with γ-TuRC and regulates the proliferative capacity of neural progenitors
Abstract
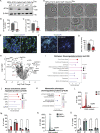
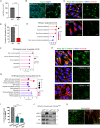
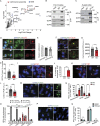
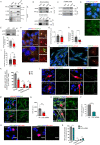
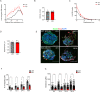
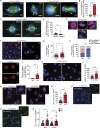
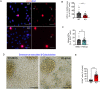
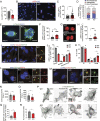
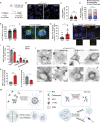
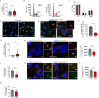
Centrosomes are organelles that nucleate microtubules via the activity of gamma-tubulin ring complexes (γ-TuRC). In the developing brain, centrosome integrity is central to the progression of the neural progenitor cell cycle, and its loss leads to microcephaly. We show that NPCs maintain centrosome integrity via the endocytic adaptor protein complex-2 (AP-2). NPCs lacking AP-2 exhibit defects in centrosome formation and mitotic progression, accompanied by DNA damage and accumulation of p53. This function of AP-2 in regulating the proliferative capacity of NPCs is independent of its role in clathrin-mediated endocytosis and is coupled to its association with the GCP2, GCP3, and GCP4 components of γ-TuRC. We find that AP-2 maintains γ-TuRC organization and regulates centrosome function at the level of MT nucleation. Taken together, our data reveal a novel, noncanonical function of AP-2 in regulating the proliferative capacity of NPCs and open new avenues for the identification of novel therapeutic strategies for the treatment of neurodevelopmental and neurodegenerative disorders with AP-2 complex dysfunction.
© 2023 Camblor-Perujo et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Andres-Alonso M, Ammar MR, Butnaru I, Gomes GM, Acuña Sanhueza G, Raman R, Yuanxiang P, Borgmeyer M, Lopez-Rojas J, Raza SA, et al. (2019) SIPA1L2 controls trafficking and local signaling of TrkB-containing amphisomes at presynaptic terminals. Nat Commun 10: 5448. 10.1038/s41467-019-13224-z - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
