Phyloecology of nitrate ammonifiers and their importance relative to denitrifiers in global terrestrial biomes
- PMID: 38086813
- PMCID: PMC10716430
- DOI: 10.1038/s41467-023-44022-3
Phyloecology of nitrate ammonifiers and their importance relative to denitrifiers in global terrestrial biomes
Abstract
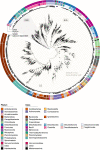
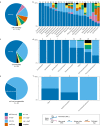
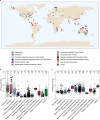
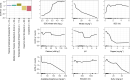
Nitrate ammonification is important for soil nitrogen retention. However, the ecology of ammonifiers and their prevalence compared with denitrifiers, being competitors for nitrate, are overlooked. Here, we screen 1 million genomes for nrfA and onr, encoding ammonifier nitrite reductases. About 40% of ammonifier assemblies carry at least one denitrification gene and show higher potential for nitrous oxide production than consumption. We then use a phylogeny-based approach to recruit gene fragments of nrfA, onr and denitrification nitrite reductase genes (nirK, nirS) in 1861 global terrestrial metagenomes. nrfA outnumbers the nearly negligible onr counts in all biomes, but denitrification genes dominate, except in tundra. Random forest modelling teases apart the influence of the soil C/N on nrfA-ammonifier vs denitrifier abundance, showing an effect of nitrate rather than carbon content. This study demonstrates the multiple roles nitrate ammonifiers play in nitrogen cycling and identifies factors ultimately controlling the fate of soil nitrate.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Kanter DR, et al. Nitrogen pollution policy beyond the farm. Nat. Food. 2020;1:27–32. doi: 10.1038/s43016-019-0001-5. - DOI
-
- Thompson RL, et al. Acceleration of global N2O emissions seen from two decades of atmospheric inversion. Nat. Clim. Change. 2019;9:993–998. doi: 10.1038/s41558-019-0613-7. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

