Panel-based RNA fusion sequencing improves diagnostics of pediatric acute myeloid leukemia
- PMID: 38086945
- PMCID: PMC10912021
- DOI: 10.1038/s41375-023-02102-9
Panel-based RNA fusion sequencing improves diagnostics of pediatric acute myeloid leukemia
Abstract
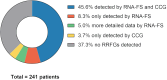
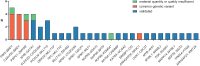
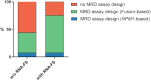
New methods like panel-based RNA fusion sequencing (RNA-FS) promise improved diagnostics in various malignancies. We here analyzed the impact of RNA-FS on the initial diagnostics of 241 cases with pediatric acute myeloid leukemia (AML). We show that, compared to classical cytogenetics (CCG), RNA-FS reliably detected risk-relevant fusion genes in pediatric AML. In addition, RNA-FS strongly improved the detection of cryptic fusion genes like NUP98::NSD1, KMT2A::MLLT10 and CBFA2T3::GLIS2 and thereby resulted in an improved risk stratification in 25 patients (10.4%). Validation of additionally detected non-risk-relevant high confidence fusion calls identified PIM3::BRD1, C22orf34::BRD1, PSPC1::ZMYM2 and ARHGAP26::NR3C1 as common genetic variants and MYB::GATA1 as recurrent aberration, which we here describe in AML subtypes M0 and M7 for the first time. However, it failed to detect rare cytogenetically confirmed fusion events like MNX1::ETV6 and other chromosome 12p-abnormalities. As add-on benefit, the proportion of patients for whom measurable residual disease (MRD) monitoring became possible was increased by RNA-FS from 44.4 to 75.5% as the information on the fusion transcripts' sequence allowed the design of new MRD assays.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Manola KN. Cytogenetics of pediatric acute myeloid leukemia. Eur J Haematol. 2009;83:391–405. - PubMed
-
- Stark B, Jeison M, Gabay LG, Mardoukh J, Luria D, Bar-Am I, et al. Classical and molecular cytogenetic abnormalities and outcome of childhood acute myeloid leukaemia: report from a referral centre in Israel. Br J Haematol. 2004;126:320–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

