Age-associated Senescent - T Cell Signaling Promotes Type 3 Immunity that Inhibits the Biomaterial Regenerative Response
- PMID: 38087458
- PMCID: PMC12522171
- DOI: 10.1002/adma.202310476
Age-associated Senescent - T Cell Signaling Promotes Type 3 Immunity that Inhibits the Biomaterial Regenerative Response
Abstract
Aging is associated with immunological changes that compromise response to infections and vaccines, exacerbate inflammatory diseases and can potentially mitigate tissue repair. Even so, age-related changes to the immune response to tissue damage and regenerative medicine therapies remain unknown. Here, it is characterized how aging induces changes in immunological signatures that inhibit tissue repair and therapeutic response to a clinical regenerative biological scaffold derived from extracellular matrix. Signatures of inflammation and interleukin (IL)-17 signaling increased with injury and treatment both locally and regionally in aged animals, and computational analysis uncovered age-associated senescent-T cell communication that promotes type 3 immunity in T cells. Local inhibition of type 3 immune activation using IL17-neutralizing antibodies improves healing and restores therapeutic response to the regenerative biomaterial, promoting muscle repair in older animals. These results provide insights into tissue immune dysregulation that occurs with aging that can be targeted to rejuvenate repair.
Keywords: aging; biomaterials; senescence; tissue engineering.
© 2023 The Authors. Advanced Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
Conflict of Interest
JHE holds equity in Unity Biotechnology and Aegeria Soft Tissue and is an advisor for Tessera Therapeutics, HapInScience, and Font Bio. DMP is consultant at Aduro Biotech, Amgen, Astra Zeneca, Bayer, Compugen, DNAtrix, Dynavax Technologies Corporation, Ervaxx, FLX Bio, Immunomic, Janssen, Merck, and Rock Springs Capital. CC is the founder and owner of C M Cherry Consulting, LLC. EJF is a member of the scientific advisory board for Viosera Therapeutics.
Figures



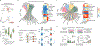

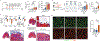
References
-
- Kirkwood TBL, Cell 2005, 120, 437. - PubMed
-
- Gosain A, Dipietro LA, World J. Surg. 2004, 28, 321. - PubMed
-
- Gould L, Abadir P, Brem H, Carter M, Conner-Kerr T, Davidson J, Dipietro L, Falanga V, Fife C, Gardner S, Grice E, Harmon J, Hazzard WR, High KP, Houghton P, Jacobson N, Kirsner RS, Kovacs EJ, Margolis D, Mcfarland Horne F, Reed MJ, Sullivan DH, Thom S, Tomic-Canic M, Walston J, Whitney JA, Williams J, Zieman S, Schmader K, J. Am. Geriatr. Soc 2015, 63, 427. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

