Deciphering Immune Landscape Remodeling Unravels the Underlying Mechanism for Synchronized Muscle and Bone Aging
- PMID: 38088531
- PMCID: PMC10837389
- DOI: 10.1002/advs.202304084
Deciphering Immune Landscape Remodeling Unravels the Underlying Mechanism for Synchronized Muscle and Bone Aging
Abstract
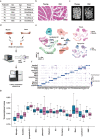
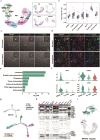
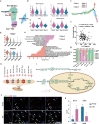
Evidence from numerous studies has revealed the synchronous progression of aging in bone and muscle; however, little is known about the underlying mechanisms. To this end, human muscles and bones are harvested and the aging-associated transcriptional dynamics of two tissues in parallel using single-cell RNA sequencing are surveyed. A subset of lipid-associated macrophages (triggering receptor expressed on myeloid cells 2, TREM2+ Macs) is identified in both aged muscle and bone. Genes responsible for muscle dystrophy and bone loss, such as secreted phosphoprotein 1 (SPP1), are also highly expressed in TREM2+ Macs, suggesting its conserved role in aging-related features. A common transition toward pro-inflammatory phenotypes in aged CD4+ T cells across tissues is also observed, activated by the nuclear factor kappa B subunit 1 (NFKB1). CD4+ T cells in aged muscle experience Th1-like differentiation, whereas, in bone, a skewing toward Th17 cells is observed. Furthermore, these results highlight that degenerated myocytes produce BAG6-containing exosomes that can communicate with Th17 cells in the bone through its receptor natural cytotoxicity triggering receptor 3 (NCR3). This communication upregulates CD6 expression in Th17 cells, which then interact with TREM2+ Macs through CD6-ALCAM signaling, ultimately stimulating the transcription of SPP1 in TREM2+ Macs. The negative correlation between serum exosomal BCL2-associated athanogene 6 (BAG6) levels and bone mineral density further supports its role in mediating muscle and bone synchronization with aging.
Keywords: aging; bone loss; muscle loss; sarcopenia; single cell analysis.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- 2020-JCJQ-QT-033/Young Elite Scientist Sponsorship Program by the China Association for Science and Technology
- 82002330,82202728,82204944/National Natural Science Foundation of China
- 2020-JQPY-003/Training Program of the National Science Fund for Distinguished Young Scholars
- 2021-JCJQ-ZQ-020/National Defense Science and Technology Fund for Distinguished Young Scholars
- 2020-JCJQ-ZD-264-2-09/Key Basic Research Projects of the Basic Strengthening Plan
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
